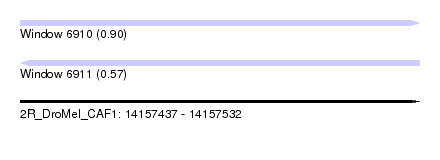
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,157,437 – 14,157,532 |
| Length | 95 |
| Max. P | 0.899484 |

| Location | 14,157,437 – 14,157,532 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
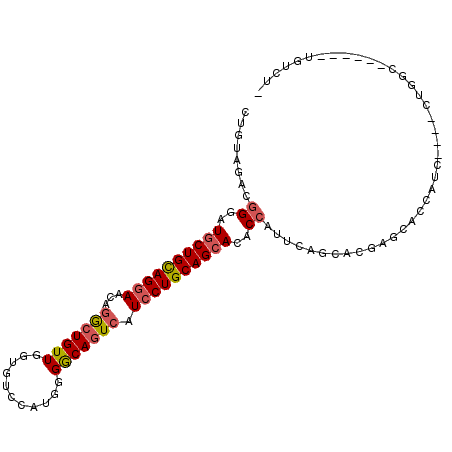
>2R_DroMel_CAF1 14157437 95 + 20766785 CUGUAGACGGGAUGCUGCAGGAACAGGCUGUUGGUGUCCAUGGGGCAGUCAUCCUGCAGCACACCAUUCAGCACGAGCACCAUC----CUGGC------UGUCUG ...(((((((..((((((((((...(((((((((...)))....)))))).))))))))))..))....(((..((......))----...))------)))))) ( -38.20) >DroVir_CAF1 3206 95 + 1 CUAUAGACAGGAUGCUGCAGGAACAGGCUGUUUGUAUCCAUCGGGCAGUCAUCCUGCAGCACACCGUUCAGCACAAGCACCAUC----CUGGU------CGUUUC .....(((((((((((((((((...(((((((((.......))))))))).)))))))))..........((....))...)))----)).))------)..... ( -38.70) >DroPse_CAF1 1869 93 + 1 CGGUAGACGGGAUGCUGCAGGAACAGGCUGUUGGUGUCCAUCGGGCAAUCAUCCUGCAGCACGCCAUUCAGCACUAGGACCAUC----CUGGC------U--CCG (((.((.(((((((((((((((...(.(((.(((...))).))).).....)))))))))..((......)).........)))----))).)------)--))) ( -36.10) >DroWil_CAF1 1235 90 + 1 CUAUACACGGGAUGCUGUAGAAACAAACUGUUGGUGUCCAUGGGGCAGUCAUCCUGCAGAACACCAUUCAAUACGAGCACCAUU----CUCCC------U----- ........(((((((((((..(((.....)))(((((.(.....((((.....)))).).)))))......))).)))).....----.))))------.----- ( -20.80) >DroMoj_CAF1 3076 105 + 1 CUGUAGACGGGAUGCUGCAGGAACAGGCUGUUCGUGUCCAUGGGGCAGUCAUCCUGCAGCACACCAUUCAGCACUAGCACCAUUGCUCUUGCUAUCCAACGUCUC ....((((((..((((((((((...((((((((.........)))))))).))))))))))..))....((((..((((....))))..)))).......)))). ( -44.90) >DroAna_CAF1 3699 94 + 1 CUGUAGACGGGAUGCUGCAGGAACAGGCUGUUGGUGUCCAUGGGACAGUCGUCCUGCAGCACUCCAUUCAAAACGAGCACCAUU----GUGGC------UGGCU- ........((..((((((((((...(((((((((...)))....)))))).))))))))))..))..........(((.((...----..)).------..)))- ( -36.80) >consensus CUGUAGACGGGAUGCUGCAGGAACAGGCUGUUGGUGUCCAUGGGGCAGUCAUCCUGCAGCACACCAUUCAGCACGAGCACCAUC____CUGGC______UGUCU_ ........((..((((((((((...(((((((...........))))))).))))))))))..))........................................ (-25.70 = -26.12 + 0.42)

| Location | 14,157,437 – 14,157,532 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -33.51 |
| Consensus MFE | -23.01 |
| Energy contribution | -22.93 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.571147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
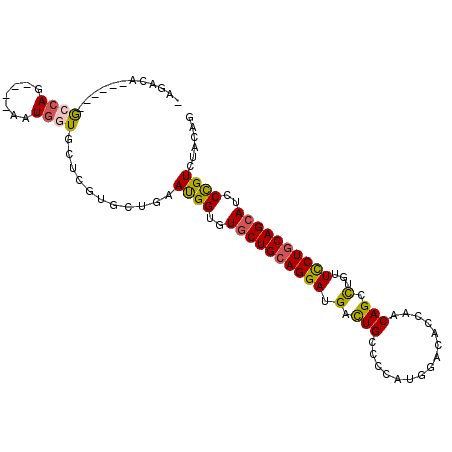
>2R_DroMel_CAF1 14157437 95 - 20766785 CAGACA------GCCAG----GAUGGUGCUCGUGCUGAAUGGUGUGCUGCAGGAUGACUGCCCCAUGGACACCAACAGCCUGUUCCUGCAGCAUCCCGUCUACAG .((((.------..(((----.(((.....))).)))...((..((((((((((.(.(((.....(((...))).))).)...))))))))))..)))))).... ( -34.90) >DroVir_CAF1 3206 95 - 1 GAAACG------ACCAG----GAUGGUGCUUGUGCUGAACGGUGUGCUGCAGGAUGACUGCCCGAUGGAUACAAACAGCCUGUUCCUGCAGCAUCCUGUCUAUAG .....(------((.((----((((.(((..(.((((....((((.(.((((.....)))).....).))))...)))).....)..))).)))))))))..... ( -30.80) >DroPse_CAF1 1869 93 - 1 CGG--A------GCCAG----GAUGGUCCUAGUGCUGAAUGGCGUGCUGCAGGAUGAUUGCCCGAUGGACACCAACAGCCUGUUCCUGCAGCAUCCCGUCUACCG (((--(------(((((----(.....))).).)))..((((.(((((((((((.....((..(.(((...))).).))....))))))))))).))))...))) ( -35.20) >DroWil_CAF1 1235 90 - 1 -----A------GGGAG----AAUGGUGCUCGUAUUGAAUGGUGUUCUGCAGGAUGACUGCCCCAUGGACACCAACAGUUUGUUUCUACAGCAUCCCGUGUAUAG -----.------((((.----..((.....((.((((..((((((((.((((.....)))).....)))))))).)))).))......))...))))........ ( -26.90) >DroMoj_CAF1 3076 105 - 1 GAGACGUUGGAUAGCAAGAGCAAUGGUGCUAGUGCUGAAUGGUGUGCUGCAGGAUGACUGCCCCAUGGACACGAACAGCCUGUUCCUGCAGCAUCCCGUCUACAG .((((......(((((..((((....))))..)))))...((..((((((((((.(.(((...............))).)...))))))))))..)))))).... ( -40.16) >DroAna_CAF1 3699 94 - 1 -AGCCA------GCCAC----AAUGGUGCUCGUUUUGAAUGGAGUGCUGCAGGACGACUGUCCCAUGGACACCAACAGCCUGUUCCUGCAGCAUCCCGUCUACAG -(((..------(((..----...)))))).((...((.(((..((((((((((((.((((....(((...)))))))..)).))))))))))..))))).)).. ( -33.10) >consensus _AGACA______GCCAG____AAUGGUGCUCGUGCUGAAUGGUGUGCUGCAGGAUGACUGCCCCAUGGACACCAACAGCCUGUUCCUGCAGCAUCCCGUCUACAG ............((((.......))))...........((((..((((((((((.(.(((...............))).)...))))))))))..))))...... (-23.01 = -22.93 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:42 2006