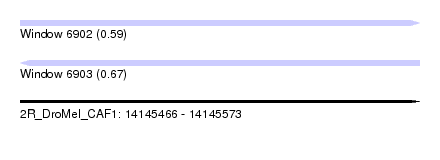
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,145,466 – 14,145,573 |
| Length | 107 |
| Max. P | 0.670592 |

| Location | 14,145,466 – 14,145,573 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -19.60 |
| Energy contribution | -21.08 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14145466 107 + 20766785 CGAUGGUUCU--GAGCCAA-CAACUUACUCUUCCUUUGCGAUACAAGGCUUUGGAGUUAUUUGGCUGGCACCGCAGCACAUGCUCCACGGACUGCAGAGCUGCGUCAUAA .((((((.((--.((((((-......(((((..(((((.....)))))....)))))...)))))))).)))(((((...(((((....))..)))..)))))))).... ( -34.30) >DroVir_CAF1 430 102 + 1 ----CGUGUU--UAACUAGC--ACUCACUCUGCCCUUGCGGUAUAAAGCCUUUGGAUUAUUUGGCUGACAACGCAAUACAUUCUCCACGGACUGUAAUGCGGCAUCAUAU ----.(((((--.....)))--))......((((.(((((......((((............)))).....)))))((((.(((....))).))))....))))...... ( -24.60) >DroGri_CAF1 400 104 + 1 ----CGUGAUAACAGCUCGC--ACUCACCCUUCCCUUGCGGUACAAUGCCUUGGGAUUAUUCGGCUGGCAUCGCAGCACAUGCUCCACGGACUGCAGUGCGGCGUCAUAG ----.(((((....(((.((--.........((((..(((......)))...)))).......)).)))...((.((((.(((((....))..))))))).))))))).. ( -31.09) >DroSec_CAF1 414 107 + 1 CUAUGGUUUU--GGGCCAA-CAACUUACUCUGCCUUUGCGGUACAAGGCUUUGGAGUUAUUUGGCUGGCACCGCAGCACAUGCUCCACGGACUGCAGAGCUGCGUCAUAA .(((((....--((((((.-(((...((((((((((........)))))...)))))...)))..)))).))(((((...(((((....))..)))..))))).))))). ( -36.60) >DroEre_CAF1 403 107 + 1 AUUUGGUGCU--GAGCUAG-CCACUUACUCUGCCCUUGCGGUACAAGGCUUUGGAGUUGUUUGGCUGGCAUCGCAGCACAUGCUCUACAGACUGUAGAGCUGCGUCAAAA .(((((((((--(.(((((-(((...((((((.(((((.....)))))...))))))....))))))))....))))))..(((((((.....))))))).....)))). ( -46.60) >DroYak_CAF1 403 107 + 1 CUCUGCUGCA--GAGCCAA-CAACUUACUCUGCCUUUGCGGUACAAGGCUUUGGAGUUAUUUGGCUGGCACCGCAGCACAUGCUCUACAGACUGCAGAGCUGCGUCAAAG ...((((((.--(.((((.-(((...((((((((((........)))))...)))))...)))..)))).).))))))((.(((((.(.....).)))))))........ ( -35.80) >consensus CU_UGGUGCU__GAGCCAA_CAACUUACUCUGCCCUUGCGGUACAAGGCUUUGGAGUUAUUUGGCUGGCACCGCAGCACAUGCUCCACGGACUGCAGAGCUGCGUCAUAA ....((((((...((((((...((((.....(((.(((.....))))))....))))...))))))))))))(((((...(((((....))..)))..)))))....... (-19.60 = -21.08 + 1.48)

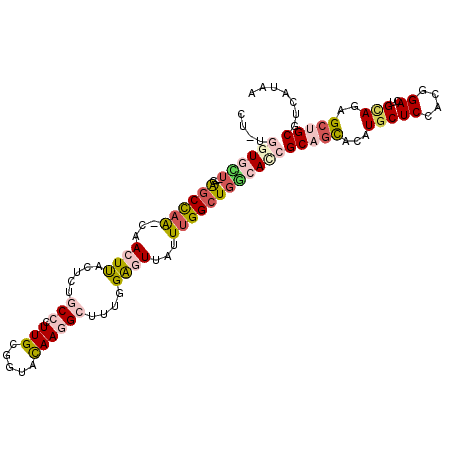
| Location | 14,145,466 – 14,145,573 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -18.45 |
| Energy contribution | -17.98 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14145466 107 - 20766785 UUAUGACGCAGCUCUGCAGUCCGUGGAGCAUGUGCUGCGGUGCCAGCCAAAUAACUCCAAAGCCUUGUAUCGCAAAGGAAGAGUAAGUUG-UUGGCUC--AGAACCAUCG .......(((((.((((..((....))))).).)))))(((...((((((.(((((......((((........)))).......)))))-)))))).--...))).... ( -32.82) >DroVir_CAF1 430 102 - 1 AUAUGAUGCCGCAUUACAGUCCGUGGAGAAUGUAUUGCGUUGUCAGCCAAAUAAUCCAAAGGCUUUAUACCGCAAGGGCAGAGUGAGU--GCUAGUUA--AACACG---- ...((((...(((((.((.((.((.((.((((.....)))).))((((............)))).....((....)))).)).)))))--))..))))--......---- ( -24.10) >DroGri_CAF1 400 104 - 1 CUAUGACGCCGCACUGCAGUCCGUGGAGCAUGUGCUGCGAUGCCAGCCGAAUAAUCCCAAGGCAUUGUACCGCAAGGGAAGGGUGAGU--GCGAGCUGUUAUCACG---- ..(((((((((((((((((..((((...))))..))))...(((.(((............)))......((....))....))).)))--))).)).)))))....---- ( -37.60) >DroSec_CAF1 414 107 - 1 UUAUGACGCAGCUCUGCAGUCCGUGGAGCAUGUGCUGCGGUGCCAGCCAAAUAACUCCAAAGCCUUGUACCGCAAAGGCAGAGUAAGUUG-UUGGCCC--AAAACCAUAG .((((..(((((.((((..((....))))).).)))))((.((((((.((...((((....(((((........))))).))))...)))-)))))))--.....)))). ( -37.80) >DroEre_CAF1 403 107 - 1 UUUUGACGCAGCUCUACAGUCUGUAGAGCAUGUGCUGCGAUGCCAGCCAAACAACUCCAAAGCCUUGUACCGCAAGGGCAGAGUAAGUGG-CUAGCUC--AGCACCAAAU .((((.....(((((((.....)))))))..((((((....((.(((((....((((....(((((........))))).))))...)))-)).)).)--))))))))). ( -41.50) >DroYak_CAF1 403 107 - 1 CUUUGACGCAGCUCUGCAGUCUGUAGAGCAUGUGCUGCGGUGCCAGCCAAAUAACUCCAAAGCCUUGUACCGCAAAGGCAGAGUAAGUUG-UUGGCUC--UGCAGCAGAG (((((.(((((((((((.....))))))).))))((((((.((((((.((...((((....(((((........))))).))))...)))-))))).)--)))))))))) ( -47.40) >consensus UUAUGACGCAGCUCUGCAGUCCGUGGAGCAUGUGCUGCGGUGCCAGCCAAAUAACUCCAAAGCCUUGUACCGCAAAGGCAGAGUAAGUUG_CUAGCUC__AGCACCA_AG .......((.(((((((.....))))))).((.((((......))))))....((((....(((((........))))).))))..........)).............. (-18.45 = -17.98 + -0.47)
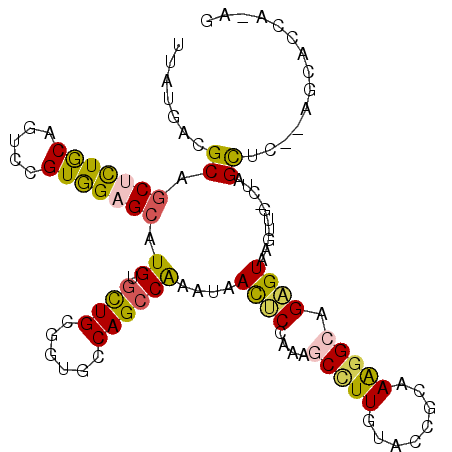
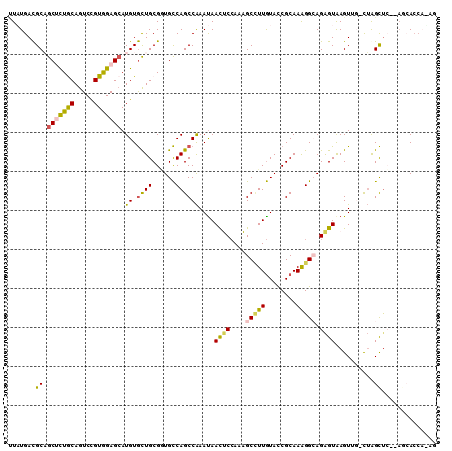
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:34 2006