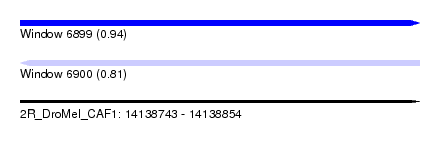
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,138,743 – 14,138,854 |
| Length | 111 |
| Max. P | 0.943721 |

| Location | 14,138,743 – 14,138,854 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -11.78 |
| Energy contribution | -12.45 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14138743 111 + 20766785 UUCUUAUCAUUUAAGU--CUAUAGCUAUUAACUUACUUGGCUGGAUCCAUGCUGUCCCUCAUGCACACCAUUCUUGGCCCCAUUUUGCCAUUUUUAUUUAGCUCCCAGCCCAC ...........(((((--..((....))..)))))...((((((.....(((((.....)).))).........((((........))))..............))))))... ( -19.40) >DroPse_CAF1 11967 101 + 1 ----------UAAAGC--CUACAAUUGGCAGCUUACUUGGCUGGAUCCAUGCUGUCCCUCAUGCAAACCAUUCGCGGACCCAUUUUCCCGUUUUUGUUCAAUUCCCAGCCAAC ----------....((--(.......))).......((((((((.....(((((.....)).)))...((...((((..........))))...))........)))))))). ( -24.70) >DroEre_CAF1 8686 111 + 1 UUCUUAUCAUUUGUGU--CUAUAGCUAUUAUCUUACUUGGCUGGAUCCAUGCUGUCCCUCAUGCUGACCAUUCUUGGGCCCAUUUUGCCAUUUUUAUUUAGCUCCCAACCCAC ...........((.((--....(((((..........(((((((..(((....(((.........)))......)))..)))....))))........)))))....)).)). ( -17.67) >DroYak_CAF1 8670 99 + 1 UUCUUAUCAAU--------------AAUUAUCUUACUUGGCUGGAUCCAUGCUGUCCCUCAUGCAGACCAUUCUCGGUCCCAUUUUGCCAUUUUUAUUUAGCUCCCAGCCCAC ...........--------------.............((((((.....(((((.....)).)))((((......)))).........................))))))... ( -19.20) >DroAna_CAF1 8878 113 + 1 UGGCUAUCAAUUAAGUAGCUUGAGCGAAGAGCUUACUUUGCCGGAUCCAUGCUGUCCCUCAUGCAGACCAUUCUGGGACCCAUUUUUCCAUUCUUAUUUGCCUCCCAGCCCAC .((((..(((.((((......(.((((((......)))))))(((...(((..(((((..(((.....)))...))))).)))...)))...)))).)))......))))... ( -26.00) >DroPer_CAF1 8643 101 + 1 ----------UAAUGC--CUACAAUUGGCAGCUUACUUGGCUGGAUCCAUGCUGUCCCUCAUGCAAACCAUUCGCGGACCCAUUUUCCCGUUUUUGUUCAAUUCCCAGCCAAC ----------...(((--(.......))))......((((((((.....(((((.....)).)))...((...((((..........))))...))........)))))))). ( -25.50) >consensus UUCUUAUCA_UUAAGU__CUACAGCUAUUAGCUUACUUGGCUGGAUCCAUGCUGUCCCUCAUGCAGACCAUUCUCGGACCCAUUUUGCCAUUUUUAUUUAGCUCCCAGCCCAC ......................................((((((.....(((((.....)).)))..((......))...........................))))))... (-11.78 = -12.45 + 0.67)

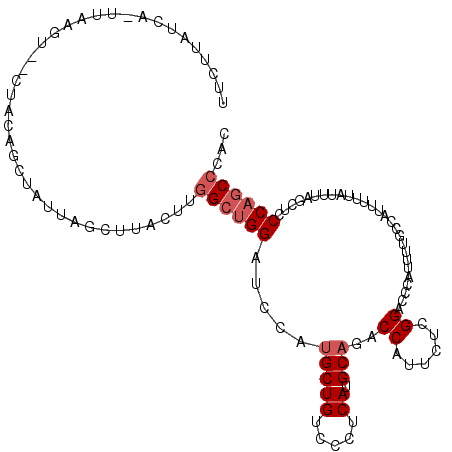
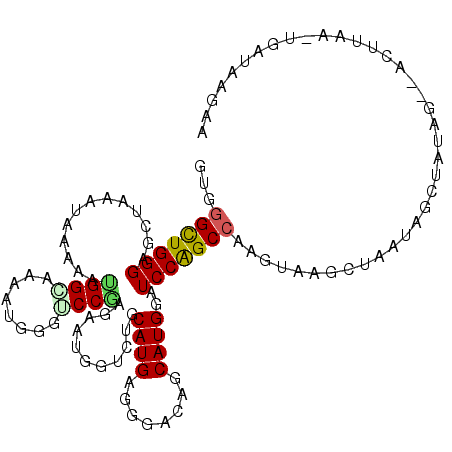
| Location | 14,138,743 – 14,138,854 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -16.43 |
| Energy contribution | -16.27 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14138743 111 - 20766785 GUGGGCUGGGAGCUAAAUAAAAAUGGCAAAAUGGGGCCAAGAAUGGUGUGCAUGAGGGACAGCAUGGAUCCAGCCAAGUAAGUUAAUAGCUAUAG--ACUUAAAUGAUAAGAA ...(((((((.............((((........))))...(((.(((.(.....).))).)))...)))))))...((((((.((....)).)--)))))........... ( -27.00) >DroPse_CAF1 11967 101 - 1 GUUGGCUGGGAAUUGAACAAAAACGGGAAAAUGGGUCCGCGAAUGGUUUGCAUGAGGGACAGCAUGGAUCCAGCCAAGUAAGCUGCCAAUUGUAG--GCUUUA---------- .(((((((((....((((.....(((((.......))).))....)))).((((........))))..)))))))))..(((((..(....)..)--))))..---------- ( -26.30) >DroEre_CAF1 8686 111 - 1 GUGGGUUGGGAGCUAAAUAAAAAUGGCAAAAUGGGCCCAAGAAUGGUCAGCAUGAGGGACAGCAUGGAUCCAGCCAAGUAAGAUAAUAGCUAUAG--ACACAAAUGAUAAGAA (((..(((..(((((........((((....(((..(((.(....(((..(....).)))..).)))..)))))))..........))))).)))--.)))............ ( -22.77) >DroYak_CAF1 8670 99 - 1 GUGGGCUGGGAGCUAAAUAAAAAUGGCAAAAUGGGACCGAGAAUGGUCUGCAUGAGGGACAGCAUGGAUCCAGCCAAGUAAGAUAAUU--------------AUUGAUAAGAA ...(((((((.((((........))))......(((((......))))).((((........))))..))))))).(((((.....))--------------)))........ ( -25.10) >DroAna_CAF1 8878 113 - 1 GUGGGCUGGGAGGCAAAUAAGAAUGGAAAAAUGGGUCCCAGAAUGGUCUGCAUGAGGGACAGCAUGGAUCCGGCAAAGUAAGCUCUUCGCUCAAGCUACUUAAUUGAUAGCCA .(((((..((((.(.........((((...(((.(((((...(((.....)))..)))))..)))...)))).........)))))..))))).(((((......).)))).. ( -31.07) >DroPer_CAF1 8643 101 - 1 GUUGGCUGGGAAUUGAACAAAAACGGGAAAAUGGGUCCGCGAAUGGUUUGCAUGAGGGACAGCAUGGAUCCAGCCAAGUAAGCUGCCAAUUGUAG--GCAUUA---------- .(((((((((....((((.....(((((.......))).))....)))).((((........))))..)))))))))......((((.......)--)))...---------- ( -27.30) >consensus GUGGGCUGGGAGCUAAAUAAAAAUGGCAAAAUGGGUCCGAGAAUGGUCUGCAUGAGGGACAGCAUGGAUCCAGCCAAGUAAGCUAAUAGCUAUAG__ACUUAA_UGAUAAGAA ...(((((((.............((((........))))...........((((........))))..)))))))...................................... (-16.43 = -16.27 + -0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:31 2006