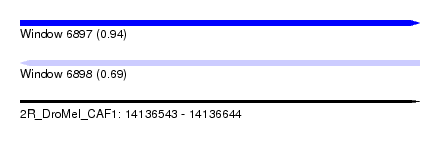
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,136,543 – 14,136,644 |
| Length | 101 |
| Max. P | 0.940876 |

| Location | 14,136,543 – 14,136,644 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -20.10 |
| Energy contribution | -21.30 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14136543 101 + 20766785 GGAGAGCCGGAGAACUCCGGACUCUCCCACGG------AACUAUAAAACACAUUUUGCGCUUGAUUGUUCAGUACAAAGGCGAUAGUUAGUUGAAUUGGUAAUUGCA--- (((((((((((....))))).)))))).....------..............(((((..((.((....))))..)))))(((((..(((((...)))))..))))).--- ( -29.90) >DroSec_CAF1 6394 101 + 1 GGAGAGCCGGAGAACUCCGGACUCUCCCACGG------AACUAUAAAAUACAUUUUGUGCUUGCUUGUUUAGUUCAAAGGCGAUAGCUAGUUCAAUUGGUAAUUGCA--- (((((((((((....))))).))))))....(------(((((......(((....((....)).))).))))))....(((((.((((((...)))))).))))).--- ( -33.50) >DroSim_CAF1 6767 101 + 1 GGAGAGCCGGAGAACUCCGGACUCUCCCACGG------AACUAUAAAACACAUUUUGUGCUUGCUUGUUCAGUGCAAAGGCGAUAGCUAGUUCAAUUGGUAAUUGCA--- (((((((((((....))))).))))))(((.(------(((....((.(((.....))).))....)))).))).....(((((.((((((...)))))).))))).--- ( -37.50) >DroEre_CAF1 6485 103 + 1 GGAGAGGCGGAGAACUCCGGACUCUCCCACGGACUCACAACUAUAAAAUACAUUUUGUGC----UUGCUCAGUGCAAAGGCAAUAGCUAGUUCAAUUGGUAAUUGCA--- ((((((.((((....))))..)))))).........................(((((..(----(.....))..)))))(((((.((((((...)))))).))))).--- ( -31.50) >DroYak_CAF1 6467 103 + 1 GGAGAGCCGGAGAACUCCGGACUCUCCCACGGGCUAUCAACUAUAAAAUACAUUUUUUGC----UUGUUCAGUGCCAAGGCAAAAGCUAGUUGAAUUGGUAAUUGCU--- (((((((((((....))))).)))))).....(((((((((((.((((......))))((----((......(((....))).)))))))))))..)))).......--- ( -34.00) >DroAna_CAF1 6678 97 + 1 GGAGUACCGGAGAACCUCGGACUCUCCCGUGGGCC--CUACUAUAAAUGA--UUU---------UCGGUCAGUGGGAAGCCGAUAGUGUGAUCGGUUGGAAGUGGUAACG .((((.((((......))))))))(((((..((((--.............--...---------..))))..)))))(((((((......)))))))............. ( -28.27) >consensus GGAGAGCCGGAGAACUCCGGACUCUCCCACGG______AACUAUAAAAUACAUUUUGUGC____UUGUUCAGUGCAAAGGCGAUAGCUAGUUCAAUUGGUAAUUGCA___ (((((((((((....))))).))))))....................................................(((((.((((((...)))))).))))).... (-20.10 = -21.30 + 1.20)


| Location | 14,136,543 – 14,136,644 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.76 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
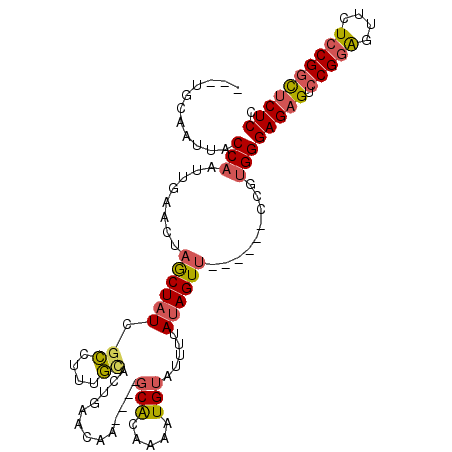
>2R_DroMel_CAF1 14136543 101 - 20766785 ---UGCAAUUACCAAUUCAACUAACUAUCGCCUUUGUACUGAACAAUCAAGCGCAAAAUGUGUUUUAUAGUU------CCGUGGGAGAGUCCGGAGUUCUCCGGCUCUCC ---.................................(((.((((.((.((((((.....)))))).)).)))------).)))((((((.(((((....))))))))))) ( -30.10) >DroSec_CAF1 6394 101 - 1 ---UGCAAUUACCAAUUGAACUAGCUAUCGCCUUUGAACUAAACAAGCAAGCACAAAAUGUAUUUUAUAGUU------CCGUGGGAGAGUCCGGAGUUCUCCGGCUCUCC ---........(((...(((((((((...((..(((.......))))).)))((.....))......)))))------)..)))(((((.(((((....)))))))))). ( -27.80) >DroSim_CAF1 6767 101 - 1 ---UGCAAUUACCAAUUGAACUAGCUAUCGCCUUUGCACUGAACAAGCAAGCACAAAAUGUGUUUUAUAGUU------CCGUGGGAGAGUCCGGAGUUCUCCGGCUCUCC ---........(((...(((((((((.((((....))...))...)))((((((.....))))))..)))))------)..)))(((((.(((((....)))))))))). ( -32.10) >DroEre_CAF1 6485 103 - 1 ---UGCAAUUACCAAUUGAACUAGCUAUUGCCUUUGCACUGAGCAA----GCACAAAAUGUAUUUUAUAGUUGUGAGUCCGUGGGAGAGUCCGGAGUUCUCCGCCUCUCC ---........(((...((.((.(((..(((....)))...)))..----.(((((.(((.....)))..)))))))))..)))(((((..((((....)))).))))). ( -24.40) >DroYak_CAF1 6467 103 - 1 ---AGCAAUUACCAAUUCAACUAGCUUUUGCCUUGGCACUGAACAA----GCAAAAAAUGUAUUUUAUAGUUGAUAGCCCGUGGGAGAGUCCGGAGUUCUCCGGCUCUCC ---.............(((((((..((((((.(((........)))----))))))...........))))))).........((((((.(((((....))))))))))) ( -30.42) >DroAna_CAF1 6678 97 - 1 CGUUACCACUUCCAACCGAUCACACUAUCGGCUUCCCACUGACCGA---------AAA--UCAUUUAUAGUAG--GGCCCACGGGAGAGUCCGAGGUUCUCCGGUACUCC ...((((.(((((..(((((......)))))...(((((((....(---------((.--...))).)))).)--)).....))))).......((....)))))).... ( -21.00) >consensus ___UGCAAUUACCAAUUGAACUAGCUAUCGCCUUUGCACUGAACAA____GCACAAAAUGUAUUUUAUAGUU______CCGUGGGAGAGUCCGGAGUUCUCCGGCUCUCC ...........(((........((((((.((....)).............(((.....))).....)))))).........)))(((((.(((((....)))))))))). (-16.34 = -16.76 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:29 2006