
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,105,383 – 14,105,488 |
| Length | 105 |
| Max. P | 0.640374 |

| Location | 14,105,383 – 14,105,488 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.60 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -9.62 |
| Energy contribution | -10.23 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14105383 105 + 20766785 AAGUAAUGGUACAAUAACAUUAAAGACUAUAACUUCAAUGUCGGUUAUUUUGGCCUGCCACACAUUUGGG-CUAUCACAGCGGGCAAUCGCCUCGCAGCCCAAAAA ......(((((.....(((((.(((.......))).))))).((((.....)))))))))....((((((-((......((((((....))).))))))))))).. ( -29.70) >DroPse_CAF1 15145 86 + 1 CAGUUAUGCCCCAAAAUUAUUGUGCAUU---------------AUGAUUUGGUGCUGCCA--CAUUUGUGCCUAUCGCAGCAUGCAAUC---UUGUAGCCCCGGCA ......((((...(((((((........---------------)))))))((.(((((((--(....)))......((.....))....---..))))).)))))) ( -19.60) >DroSec_CAF1 14609 94 + 1 AAGUAAUGAUCCAAUAACA---------AUAACUUCA--GUCGCUUAUUUUGGCCUGCCACACAUUUGGG-CUAUCACAGCGGGCAAUCGCCUCGCAGCCCAAAAA ((((..(((..........---------......)))--...))))....(((....)))....((((((-((......((((((....))).))))))))))).. ( -23.69) >DroSim_CAF1 27832 94 + 1 AAGUAAUGAUCCAAUAACA---------AUAACUUCA--GUCGCUUAUUUUGGCCUGCCACACAUUUGGG-CUAUCACAGCAGGCAAUCGCCUCGCAGCCCAAAAA ((((..(((..........---------......)))--...))))....(((....)))....((((((-((......((((((....)))).)))))))))).. ( -22.19) >DroEre_CAF1 9752 102 + 1 ACGUAAUGGUCCAAUAACAUUUAAGAUUACAAAUUCA--GUCGGUUAUUUCGGCCUGCCACACAUUUGGG-CUAUCACAGCGGGCAAUCGCCUCGCAGUCUCA-AA ..((.(((((((((..........((........)).--(((((.....)))))...........)))))-)))).)).((((((....))).))).......-.. ( -26.70) >DroYak_CAF1 25610 103 + 1 ACGUAAUGGUCCAAUAACAUAAAAGAUUAUAACUUCA--GUCGAUUAUUUCGAAGUGCCACACAUUUGGG-CUAUCACGGCGGGCAAUCGCCUCGCAGUGCCAAAA .(((.(((((((((.......................--.((((.....)))).(((...)))..)))))-)))).)))((((((....))).))).......... ( -28.20) >consensus AAGUAAUGGUCCAAUAACAUU_AAGAUUAUAACUUCA__GUCGAUUAUUUUGGCCUGCCACACAUUUGGG_CUAUCACAGCGGGCAAUCGCCUCGCAGCCCAAAAA ...............................................((((((.((((.......(((.........))).((((....)))).)))).)))))). ( -9.62 = -10.23 + 0.62)

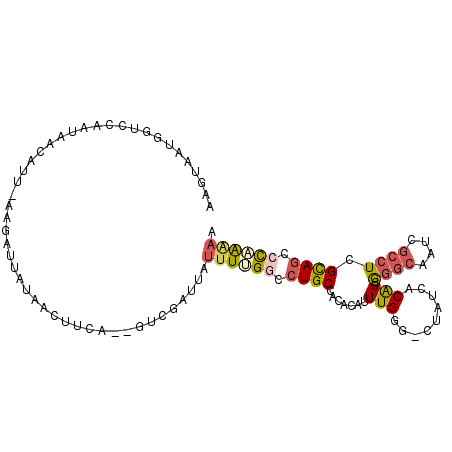

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:24 2006