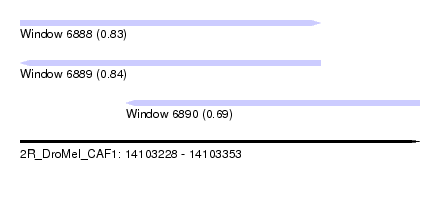
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,103,228 – 14,103,353 |
| Length | 125 |
| Max. P | 0.837357 |

| Location | 14,103,228 – 14,103,322 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -29.26 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
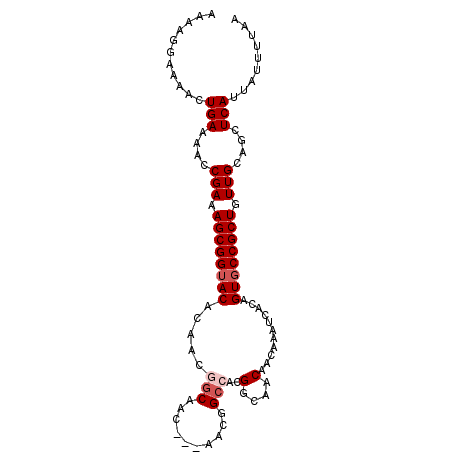
>2R_DroMel_CAF1 14103228 94 + 20766785 GACAGUCGGUAGAAAGUGCCGCCGGCGGCACCGUUAAAAUAAUGAGCUGCAACAGCGGCACUGUGAUUUGUUGUUGCCGUGGCCGUU------GUCGUUG (((((.((((....((((((((..(((((..(((((...))))).)))))....))))))))(..((.....))..)....))))))------))).... ( -37.60) >DroSec_CAF1 12305 94 + 1 GACAGUCGGUAGAAAGUGCCGCCGGCGGCACCGUUAAAAUAAUGAGCUGCAACAGCGGCACUGUGAUUUGUUGUUGCCGUGGCCGUU------GCCGUUG ..(((.((((((...(.(((((.(((((((.(((((...))))).(((((....)))))............))))))))))))).))------))))))) ( -36.30) >DroSim_CAF1 25577 100 + 1 GACAGUCGGUAGAAAGUGCCGCCGGCGGCACCGUUAAAAUAAUGAGCUGCAACAGCGGCACUGUGAUUUGUUGUUGCCGUGGCCGUUGCCGUUGCCGUUG (.(((.((((((...(.(((((.(((((((.(((((...))))).(((((....)))))............))))))))))))).))))))))).).... ( -37.10) >DroEre_CAF1 7480 94 + 1 GACAGUCGGUAGAAAGUGCCGCCGGCGGCACCGUUAAAAUAAUGAGCUGCAACAGCGGAACUGUGAUUUGUUGUUGCCAUUGC------CGUUGCCGUUG ..(((.((((.((..((((((....))))))..)).....((((.((.(((((((((((.......)))))))))))....))------))))))))))) ( -33.00) >DroYak_CAF1 23191 100 + 1 GACAGUCGGUAGAAAGUGCCGCCGGCGGCACCGUUAAAAUAAUGAGCUGCAACAGCGGCACUGUGAUUUGUUGUUGCCGCUGCCGUUGCUGUUGCCGUUG ((((((((((((..((((((((..(((((..(((((...))))).)))))....))))))))(..((.....))..)..))))))..))))))....... ( -40.80) >consensus GACAGUCGGUAGAAAGUGCCGCCGGCGGCACCGUUAAAAUAAUGAGCUGCAACAGCGGCACUGUGAUUUGUUGUUGCCGUGGCCGUU___GUUGCCGUUG ..(((.((((....((((((((..(((((..(((((...))))).)))))....))))))))(..((.....))..)................))))))) (-29.26 = -29.30 + 0.04)

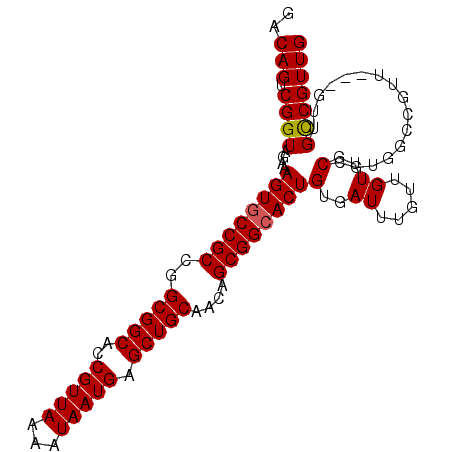
| Location | 14,103,228 – 14,103,322 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -24.08 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14103228 94 - 20766785 CAACGAC------AACGGCCACGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAACGGUGCCGCCGGCGGCACUUUCUACCGACUGUC ....(((------(.(((....(....)..........((((((((((..(((.(.............).)))...)))))))))).....)))..)))) ( -28.42) >DroSec_CAF1 12305 94 - 1 CAACGGC------AACGGCCACGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAACGGUGCCGCCGGCGGCACUUUCUACCGACUGUC ...((((------((((((...((((..............))))))))))))................(((((((....)))))))......))...... ( -29.44) >DroSim_CAF1 25577 100 - 1 CAACGGCAACGGCAACGGCCACGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAACGGUGCCGCCGGCGGCACUUUCUACCGACUGUC ...(((.....((((((((...((((..............))))))))))))................(((((((....))))))).....)))...... ( -32.04) >DroEre_CAF1 7480 94 - 1 CAACGGCAACG------GCAAUGGCAACAACAAAUCACAGUUCCGCUGUUGCAGCUCAUUAUUUUAACGGUGCCGCCGGCGGCACUUUCUACCGACUGUC ...((((((((------((..((....))((........))...))))))))................(((((((....)))))))......))...... ( -27.80) >DroYak_CAF1 23191 100 - 1 CAACGGCAACAGCAACGGCAGCGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAACGGUGCCGCCGGCGGCACUUUCUACCGACUGUC ...(((....(((..(((((((((((..............)))))))))))..)))............(((((((....))))))).....)))...... ( -36.74) >consensus CAACGGCAAC___AACGGCCACGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAACGGUGCCGCCGGCGGCACUUUCUACCGACUGUC ...(((...........((....)).............((((((((((..(((.(.............).)))...)))))))))).....)))...... (-24.08 = -24.48 + 0.40)


| Location | 14,103,261 – 14,103,353 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -15.92 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14103261 92 - 20766785 AAAAGGAAAACUGAAAACCGAAAGCGGUACACAACGAC------AACGGCCACGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAA ...........(((....(((.((((((((....((..------..))((....))..............)))))))).)))....)))......... ( -19.00) >DroSec_CAF1 12338 92 - 1 AAAAGGAAAACUGAAAACCGAAAGCGGUACACAACGGC------AACGGCCACGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAA ...........(((....(((.((((((((....((..------..))((....))..............)))))))).)))....)))......... ( -22.80) >DroSim_CAF1 25610 98 - 1 AAAAGGAAAACUGAAAACCGAAAGCGGUACACAACGGCAACGGCAACGGCCACGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAA ...........(((....(((.((((((((.....(((..((....)))))..(....)...........)))))))).)))....)))......... ( -25.80) >DroEre_CAF1 7513 92 - 1 AAAAGGAAAACUGAAAAUCGAAAGCGGUACACAACGGCAACG------GCAAUGGCAACAACAAAUCACAGUUCCGCUGUUGCAGCUCAUUAUUUUAA ...........(((....(((.(((((.((....((....))------....((....))..........)).))))).)))....)))......... ( -17.80) >DroYak_CAF1 23224 98 - 1 AAAAGGAAAACUGAAAAUCGAAAGCGGUACACAACGGCAACAGCAACGGCAGCGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAA ...........(((((.......((.((.....)).))...(((..(((((((((((..............)))))))))))..))).....))))). ( -25.24) >consensus AAAAGGAAAACUGAAAACCGAAAGCGGUACACAACGGCAAC___AACGGCCACGGCAACAACAAAUCACAGUGCCGCUGUUGCAGCUCAUUAUUUUAA ...........(((....(((.((((((((.....(((..........)))..(....)...........)))))))).)))....)))......... (-15.92 = -16.72 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:22 2006