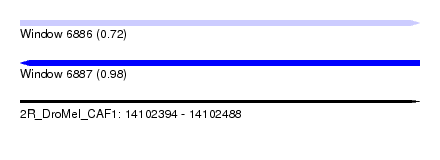
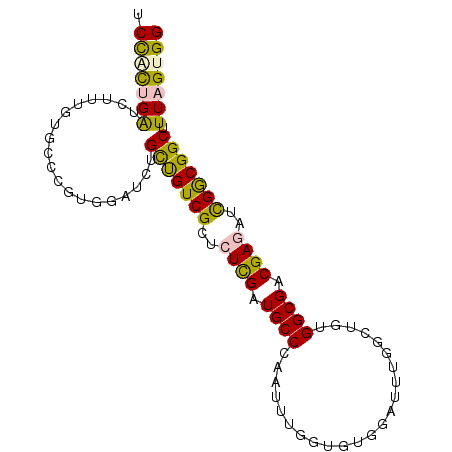
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,102,394 – 14,102,488 |
| Length | 94 |
| Max. P | 0.982037 |

| Location | 14,102,394 – 14,102,488 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -8.84 |
| Energy contribution | -8.85 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14102394 94 + 20766785 CCACUAAAGCCGCCAAUCUCGUCGCCACAGCCAAAUCCACACCAAAUUGGGCAUCGAGAGCGACAGCAGAUCCACGGGCACAAAGAUCAGUGGA (((((...(((((...(((((..(((.(((................))))))..))))))))...)).((((...(....)...))))))))). ( -24.79) >DroPse_CAF1 12701 77 + 1 CCACAAAUGCCGUCGAGAUCGUCGCCAAAGCC-----------AAAUUGGGCAUCAAGAGCGACGACAUCGACA------UCGACAUCGAGAGA ...........(((((..((((((((...(((-----------......))).....).)))))))..))))).------(((....))).... ( -26.10) >DroSec_CAF1 11422 94 + 1 CCACUAAAGCCGCCAAUCUCGUCGCCACAACCAAAUCCACACCAAAUUGGGCAUCGAGAGCGACAGCAGAUCCACGGGCACAAAGAUCAGUGGA (((((...(((((...(((((..(((.(((................))))))..))))))))...)).((((...(....)...))))))))). ( -25.09) >DroSim_CAF1 24694 94 + 1 CCACUAAAGCCGCCAAUCUCGUCGCCACAACCAAAUCCACACCAAAUUGGGCAUCGAGAGCGACAGCAGAUCCACGGACACAAAGAUCAGUGGA (((((...(((((...(((((..(((.(((................))))))..))))))))...)).((((...(....)...))))))))). ( -25.09) >DroEre_CAF1 6614 94 + 1 CCACUAAAGCCGCCGUUCUCGUCGCCACAGCCAAAUCCAAACAAAAUUGGGCAUCAAGUGCGACAGCAGAUCCACGGGCACAAAGACCAGCGGA .........((((.(.(((.(((((.((.......(((((......)))))......))))))).((.(.....)..))....))).).)))). ( -20.72) >DroPer_CAF1 12621 83 + 1 CCACAAAUGCCGUCGAGAUCGUCGCCAAAGCC-----------AAAUUGGGCAUCAAGAGCGACGACAUCGACAUCGACAUCGACAUCGAGAGA ...........(((((..((((((((...(((-----------......))).....).)))))))..))))).(((....)))..((....)) ( -26.60) >consensus CCACUAAAGCCGCCAAUCUCGUCGCCACAGCCAAAUCCACACCAAAUUGGGCAUCAAGAGCGACAGCAGAUCCACGGGCACAAAGAUCAGUGGA .........(((....(((.((((((...(((.................))).....).)))))...)))....)))................. ( -8.84 = -8.85 + 0.00)

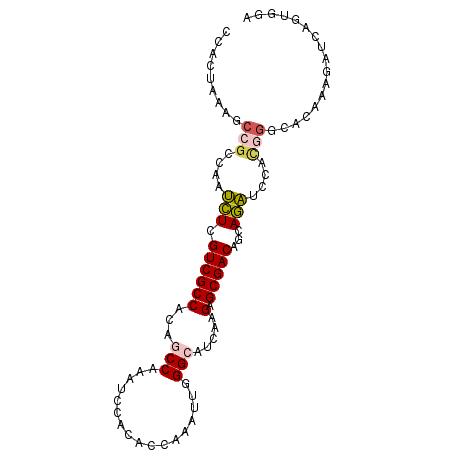
| Location | 14,102,394 – 14,102,488 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.07 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14102394 94 - 20766785 UCCACUGAUCUUUGUGCCCGUGGAUCUGCUGUCGCUCUCGAUGCCCAAUUUGGUGUGGAUUUGGCUGUGGCGACGAGAUUGGCGGCUUUAGUGG .((((((((((.((....)).))))).(((((((.(((((.((((((.((..((....))..)).)).)))).))))).)))))))...))))) ( -34.00) >DroPse_CAF1 12701 77 - 1 UCUCUCGAUGUCGA------UGUCGAUGUCGUCGCUCUUGAUGCCCAAUUU-----------GGCUUUGGCGACGAUCUCGACGGCAUUUGUGG ......((((((..------.(((((.(((((((((...((.(((......-----------))))).))))))))).)))))))))))..... ( -29.90) >DroSec_CAF1 11422 94 - 1 UCCACUGAUCUUUGUGCCCGUGGAUCUGCUGUCGCUCUCGAUGCCCAAUUUGGUGUGGAUUUGGUUGUGGCGACGAGAUUGGCGGCUUUAGUGG .((((((((((.((....)).))))).(((((((.(((((.(((((((((..((....))..))))).)))).))))).)))))))...))))) ( -38.00) >DroSim_CAF1 24694 94 - 1 UCCACUGAUCUUUGUGUCCGUGGAUCUGCUGUCGCUCUCGAUGCCCAAUUUGGUGUGGAUUUGGUUGUGGCGACGAGAUUGGCGGCUUUAGUGG .((((((((((.((....)).))))).(((((((.(((((.(((((((((..((....))..))))).)))).))))).)))))))...))))) ( -38.00) >DroEre_CAF1 6614 94 - 1 UCCGCUGGUCUUUGUGCCCGUGGAUCUGCUGUCGCACUUGAUGCCCAAUUUUGUUUGGAUUUGGCUGUGGCGACGAGAACGGCGGCUUUAGUGG (((((.(((......))).)))))...(((((((..((((.((((((.((..((....))..)).)).)))).))))..)))))))........ ( -30.40) >DroPer_CAF1 12621 83 - 1 UCUCUCGAUGUCGAUGUCGAUGUCGAUGUCGUCGCUCUUGAUGCCCAAUUU-----------GGCUUUGGCGACGAUCUCGACGGCAUUUGUGG ....(((((......)))))((((((.(((((((((...((.(((......-----------))))).))))))))).)))))).......... ( -30.80) >consensus UCCACUGAUCUUUGUGCCCGUGGAUCUGCUGUCGCUCUCGAUGCCCAAUUUGGUGUGGAUUUGGCUGUGGCGACGAGAUCGGCGGCUUUAGUGG .(((((((...................(((((((..((((.((((.......................)))).))))..))))))).))))))) (-19.90 = -19.07 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:19 2006