
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,100,347 – 14,100,459 |
| Length | 112 |
| Max. P | 0.728902 |

| Location | 14,100,347 – 14,100,459 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -30.78 |
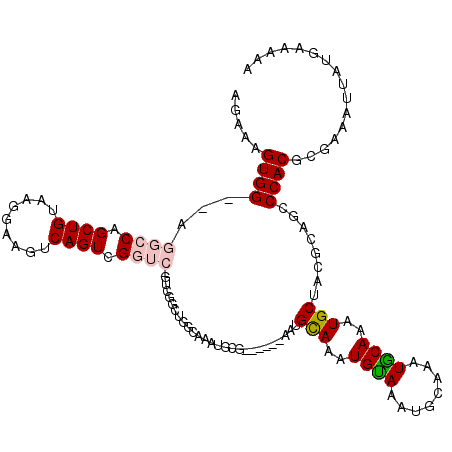
| Consensus MFE | -14.93 |
| Energy contribution | -15.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
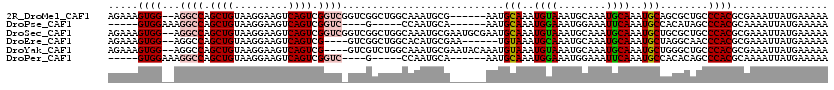
>2R_DroMel_CAF1 14100347 112 + 20766785 AGAAAGUGG--AGGCCAGCUGUAAGGAAGUCAGUCGGUCGGUCGGCUGGCAAAUGCG------AAUGCAAAUGUAAAUGCAAAUGCAAAUGCAGCGCUGCCCACGCGAAAUUAUGAAAAA .....(((.--.(((..((((((.....(((((((((....)))))))))...((((------..((((........))))..))))..))))))...)))..))).............. ( -38.90) >DroPse_CAF1 10363 100 + 1 -----GUGGAAAGGCCAGCUGUAAGGAAGUCAGUCGGUC----G-----CCAAUGCA------AAUGCAAAUGGAAAUGGAAAUUCAAAUGCCACAUAGCCCACGCAAAAUUAUGAAAAA -----((((...((((.((((.........)))).))))----(-----(...(((.------...)))..(((.....(.....).....)))....))))))................ ( -21.10) >DroSec_CAF1 9381 118 + 1 AGAAAGUGG--AGGCCAGCUGUAAGGAAGUCAGUCGGUCGGUCGGCUGGCAAAUGCGAAUGCGAAUGCAAAUGUAAAUGCAAAUGCAAAUGCUGCGCUGCCCACGCGAAAUUAUGAAAAA .....((((--.(((((((.........(((((((((....)))))))))...((((..((((..(((....)))..))))..))))...)))).)))..))))................ ( -38.60) >DroEre_CAF1 4633 108 + 1 AGAAAGUGG--AGGCCAGCUGUAAGGAAGUCAGUCG----GUCGGCUGGCACAUGCGAA------UGUAAAUGCAAAUGCAAAUGCAAAUGCUAGGCAACCCACGCGAAAUUAUGAAAAA .....((((--.((((.((((.........)))).)----)))(.((((((..((((..------((((........))))..))))..)))))).)...))))................ ( -32.90) >DroYak_CAF1 20007 114 + 1 AGAAAGUGG--AGGCCAGCUGUAAGGAAGUCAGUCG----GUCGUCUGGCAAAUGCGAAUACAAAUGUAAAUGUAAAUGCAAAUGCAAAUGCUGGGCUGCCCACGCGAAAUUAUGAAAAA .....((((--.((((.((((.........)))).)----)))(((..(((..((((..(((....)))..))))..(((....)))..)))..)))...))))................ ( -32.10) >DroPer_CAF1 10303 100 + 1 -----GUGGAAAGGCCAGCUGUAAGGAAGUCAGUCGGUC----G-----CCAAUGCA------AAUGCAAAUGGAAAUGGAAAUUCAAAUGCCACACAGCCCACGCAAAAUUAUGAAAAA -----((((...((((.((((.........)))).))))----(-----(...(((.------...)))..(((.....(.....).....)))....))))))................ ( -21.10) >consensus AGAAAGUGG__AGGCCAGCUGUAAGGAAGUCAGUCGGUC_GUCGGCUGGCAAAUGCG______AAUGCAAAUGUAAAUGCAAAUGCAAAUGCUACGCAGCCCACGCGAAAUUAUGAAAAA .....((((...((((.((((.........)))).))))...........................(((..((((........))))..)))........))))................ (-14.93 = -15.18 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:17 2006