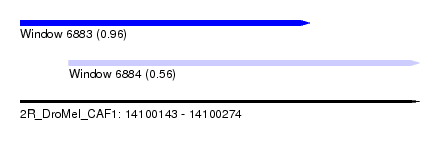
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,100,143 – 14,100,274 |
| Length | 131 |
| Max. P | 0.963639 |

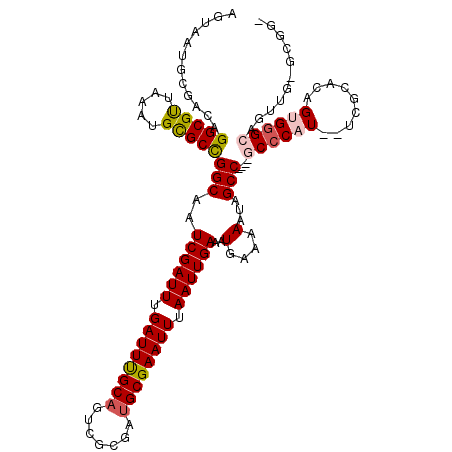
| Location | 14,100,143 – 14,100,238 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.59 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.69 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
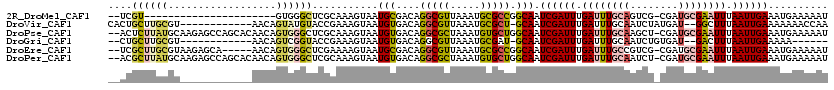
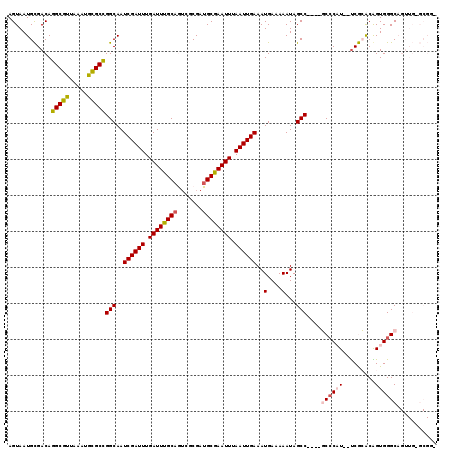
>2R_DroMel_CAF1 14100143 95 + 20766785 --UCGU----------------------GUGGGCUCGCAAAGUAAUGCGACAGGCGUUAAAUGCGCCGGCAAUCGAUUUGAUUUGCAGUCG-CGAUGCGAAUUUAAUUGAAAUGAAAAAU --((((----------------------((.(((..(((...((((((.....))))))..)))))).))..((((((.((((((((....-...)))))))).)))))).))))..... ( -30.50) >DroVir_CAF1 14147 105 + 1 CACUGCUUGCGU------------AACAGUAUGUACCGAAAGUAAUGUGACAGGCGUUAAAUGCGCU-GCAAUCGAUUUGAUUUGCAAUCUAUGAU--GGCUUUAAUUGAAAAAAACCAA ...(((..((((------------(...........(....)((((((.....))))))..))))).-))).((((((......((.(((...)))--.))...)))))).......... ( -17.80) >DroPse_CAF1 10170 117 + 1 --ACUCUUAUGCAAGAGCCAGCACAACAGUGGGCUCGCAAAGUAAUGUGACAGGCGCUAAAUGUGCUGGCAAUCGAUUUGAUUUGCAAGCU-CGAUGCGAAUUUAAUUGAAAUGAAAAAU --..((((....))))(((((((((..((((.(.(((((......))))))...))))...)))))))))..((((((.((((((((....-...)))))))).)))))).......... ( -37.40) >DroGri_CAF1 14403 97 + 1 --CUGCUUGCGU------------AACAGUCGGUACCGAAAGUAAUGUGACAGGCGUUAAAUGCGAU-GCAAUCGAUUUGAUUUGCAAUCUGUGAU--GACUUUAAUUGAAAAA------ --(((.(..(((------------.((..(((....)))..)).)))..))))(((((......)))-))..((((((....(..(.....)..).--......))))))....------ ( -20.20) >DroEre_CAF1 4415 112 + 1 --UCGCUUGCGUAAGAGCA-----AACAGUGGGCUCGAAAAGUAAUGCGACAGGCGUUAAAUGCGCCGGCAAUCGAUUUGAUUUGCCGUCG-CGAUGCGAAUUUAAUUGAAAUGAAAAAU --((((((((....((((.-----........)))).....))))((((((.(((((.....)))))(((((((.....)).)))))))))-))..)))).................... ( -37.30) >DroPer_CAF1 10117 117 + 1 --ACGCUUAUGCAAGAGCCAGCACAACAGUGGGCUCGCAAAGUAAUGUGACAGGCGCUAAAUGUGCUGGCAAUCGAUUUGAUUUGCAAUCU-CGAUGCGAAUUUAAUUGAAAUGAAAAAU --..((....))....(((((((((..((((.(.(((((......))))))...))))...)))))))))..((((((.((((((((....-...)))))))).)))))).......... ( -38.70) >consensus __ACGCUUGCGU____________AACAGUGGGCUCGCAAAGUAAUGUGACAGGCGUUAAAUGCGCUGGCAAUCGAUUUGAUUUGCAAUCU_CGAUGCGAAUUUAAUUGAAAUGAAAAAU ....((((((..................))))))...........(((....(((((.....))))).))).((((((.((((((((........)))))))).)))))).......... (-16.93 = -17.69 + 0.75)
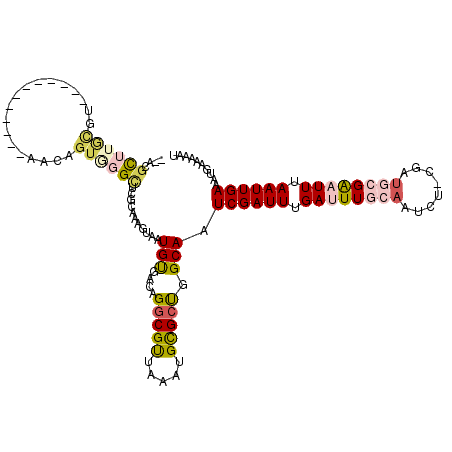
| Location | 14,100,159 – 14,100,274 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -26.89 |
| Energy contribution | -27.08 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14100159 115 + 20766785 AGUAAUGCGACAGGCGUUAAAUGCGCCGGCAAUCGAUUUGAUUUGCAGUCGCGAUGCGAAUUUAAUUGAAAUGAAAAAUAGCC----GCCCAUAGCCGCACGGUGGGCAGUUGUGCGCC ......(((((((((((.....)))))(((..((((((.((((((((.......)))))))).))))))..(....)...)))----((((...(((....)))))))...))).))). ( -40.80) >DroPse_CAF1 10208 106 + 1 AGUAAUGUGACAGGCGCUAAAUGUGCUGGCAAUCGAUUUGAUUUGCAAGCUCGAUGCGAAUUUAAUUGAAAUGAAAAAUAGCCAGCCCCCCUU--UCAGGCAGUGGGC----------- ............(((((.....)))))(((..((((((.((((((((.......)))))))).))))))..(....)...))).(((((((..--...))..).))))----------- ( -28.10) >DroSim_CAF1 22437 113 + 1 AGUAAUGCGACAGGCGUUAAAUGCGCCGGCAAUCGAUUUGAUUUGCAGUCGCGAUGCGAAUUUAAUUGAAAUGAAAAAUAGCC----GCCCAU--UCGCACGGUGGGCAGUUGUGCGGC .((..((((((.(((((.....)))))(((..((((((.((((((((.......)))))))).))))))..(....)...)))----((((((--(.....))))))).))))))..)) ( -41.00) >DroEre_CAF1 4448 112 + 1 AGUAAUGCGACAGGCGUUAAAUGCGCCGGCAAUCGAUUUGAUUUGCCGUCGCGAUGCGAAUUUAAUUGAAAUGAAAAAUAGCC----GCGCAU--UCGCGCGGUGGGCAGUUG-GCGGU .(((.((((((.(((((.....)))))(((((((.....)).))))))))))).)))....(((((((............(((----((((..--..)))))))...))))))-).... ( -44.06) >DroYak_CAF1 19811 112 + 1 AGUAAUGCGACAGGCGUUAAAUGCGCCGGCAAUCGAUUUGAUUCGCAGUCGCGAUGCGAAUUUAAUUGAAAUGAAAAAUAGCC----GCCCAU--UCGCACAGUGGGCAGUUG-GCGGU ..((((((.....))))))....(((((((..((((((.((((((((.......)))))))).))))))..............----((((((--(.....))))))).))))-))).. ( -42.60) >DroPer_CAF1 10155 106 + 1 AGUAAUGUGACAGGCGCUAAAUGUGCUGGCAAUCGAUUUGAUUUGCAAUCUCGAUGCGAAUUUAAUUGAAAUGAAAAAUAGCCAGCCCCCCUU--UCAGGCAGUGGGC----------- ............(((((.....)))))(((..((((((.((((((((.......)))))))).))))))..(....)...))).(((((((..--...))..).))))----------- ( -28.10) >consensus AGUAAUGCGACAGGCGUUAAAUGCGCCGGCAAUCGAUUUGAUUUGCAGUCGCGAUGCGAAUUUAAUUGAAAUGAAAAAUAGCC____GCCCAU__UCGCACAGUGGGCAGUUG_GCGG_ ............(((((.....)))))(((..((((((.((((((((.......)))))))).)))))).((.....)).)))....((((((.........))))))........... (-26.89 = -27.08 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:16 2006