
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,088,610 – 14,088,725 |
| Length | 115 |
| Max. P | 0.903810 |

| Location | 14,088,610 – 14,088,725 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -24.79 |
| Energy contribution | -24.13 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
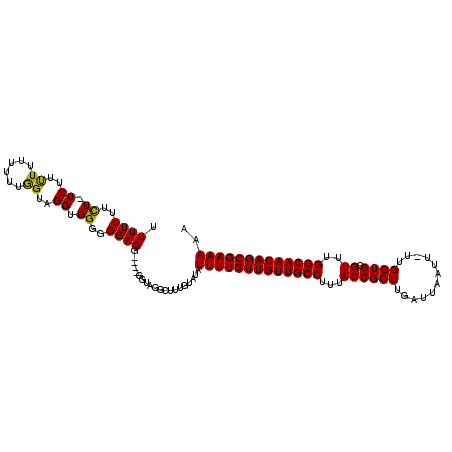
>2R_DroMel_CAF1 14088610 115 + 20766785 UUUUUUUCUCUCUUUUUUUUGUAGUUGGAGGGGGGAGGAGUAGGUAGGCUUUGUAUAUUUUGUUGUUGCCUUUACCGCUUGAUUAAUUUUUGGUGCGUUUGGCAACAGCGAAAAA (((((..((((((..((.....))..))))))..)))))..................(((((((((((((...((((((.((......)).)))).))..))))))))))))).. ( -36.30) >DroSec_CAF1 3806 108 + 1 UUUUUUUUU-UC-UCUUUUUUUGGUAGGUGGGGGGAGG----GGUAGGCUUUGUAUAUUUUGUUGUUGCCUUUACCGCUUGAUUAAUU-UUGGUGCGUUUGGCAACAGCGAAAAA .......(.-.(-(((((((.........)))))))).----.).............(((((((((((((...((((((.((.....)-).)))).))..))))))))))))).. ( -30.20) >DroSim_CAF1 13476 110 + 1 UUUUUUUCUAUCUUUUUUUUUUGGUAGGUGGGGGGAGG----GGUAGGCUUUGUAUAUUUUGUUGUUGCCUUUACCGCUUGAUUAAUU-UUGGUGCGUUUGGCAACAGCGAAAAA ......(((((((((((((((........)))))))))----)))))).........(((((((((((((...((((((.((.....)-).)))).))..))))))))))))).. ( -33.60) >consensus UUUUUUUCU_UCUUUUUUUUUUGGUAGGUGGGGGGAGG____GGUAGGCUUUGUAUAUUUUGUUGUUGCCUUUACCGCUUGAUUAAUU_UUGGUGCGUUUGGCAACAGCGAAAAA .((((..((.((..((......))..)).))..))))....................(((((((((((((...((((((............)))).))..))))))))))))).. (-24.79 = -24.13 + -0.66)

| Location | 14,088,610 – 14,088,725 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -14.92 |
| Consensus MFE | -12.99 |
| Energy contribution | -12.99 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
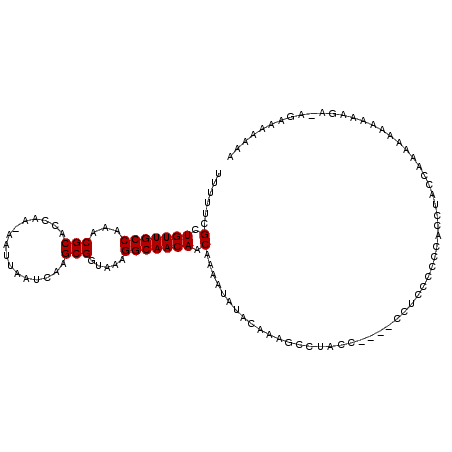
>2R_DroMel_CAF1 14088610 115 - 20766785 UUUUUCGCUGUUGCCAAACGCACCAAAAAUUAAUCAAGCGGUAAAGGCAACAACAAAAUAUACAAAGCCUACCUACUCCUCCCCCCUCCAACUACAAAAAAAAGAGAGAAAAAAA (((((((.(((((((...(((................))).....))))))).)...............................(((...............))).)))))).. ( -16.55) >DroSec_CAF1 3806 108 - 1 UUUUUCGCUGUUGCCAAACGCACCAA-AAUUAAUCAAGCGGUAAAGGCAACAACAAAAUAUACAAAGCCUACC----CCUCCCCCCACCUACCAAAAAAAGA-GA-AAAAAAAAA (((((((.(((((((...(((.....-..........))).....))))))).)...................----...........((.........)).-))-))))..... ( -14.26) >DroSim_CAF1 13476 110 - 1 UUUUUCGCUGUUGCCAAACGCACCAA-AAUUAAUCAAGCGGUAAAGGCAACAACAAAAUAUACAAAGCCUACC----CCUCCCCCCACCUACCAAAAAAAAAAGAUAGAAAAAAA (((((((.(((((((...(((.....-..........))).....))))))).)...................----...........((............))...)))))).. ( -13.96) >consensus UUUUUCGCUGUUGCCAAACGCACCAA_AAUUAAUCAAGCGGUAAAGGCAACAACAAAAUAUACAAAGCCUACC____CCUCCCCCCACCUACCAAAAAAAAAAGA_AGAAAAAAA ......(.(((((((...(((................))).....))))))).)............................................................. (-12.99 = -12.99 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:11 2006