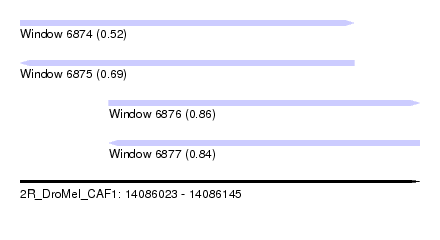
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,086,023 – 14,086,145 |
| Length | 122 |
| Max. P | 0.858223 |

| Location | 14,086,023 – 14,086,125 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -16.04 |
| Energy contribution | -15.85 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
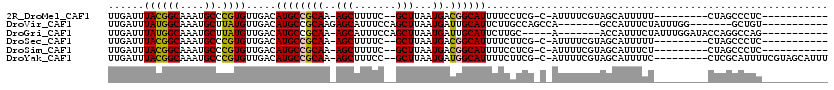
>2R_DroMel_CAF1 14086023 102 + 20766785 UGUUGGGAUCUCUAGAACU---UGC-UGCAUUUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA-AGCUUUUC--GCUUAAUGACGGCAUUUUCCUCG-C-AUUUUC (((.((((.......(((.---((.-.(((((((........))))))).)).)))...(((((((((-(((.....--))))..)).))))))..)))).)-)-)..... ( -26.80) >DroVir_CAF1 2089 103 + 1 UGUUGGCAU---UAAUGCUUUUGGCCGUCAUUUGAUUUAUGGCAAAUGCUUAUGUUGACAUGCCGCAAGAGCAUUUCCAGCUUAAUGAUUGCAUUCUUGCCAGCCA----- .(((((((.---.(((((..(((((.(((....)))...(((.((((((((.(((.(.....).))).))))))))))))))....))..)))))..)))))))..----- ( -33.20) >DroGri_CAF1 2320 97 + 1 UGUUGGCAU---UAAUGCUUUUGGCCGUCAUUUGAUUUAUGGCAAAUGCUUAUGUUGACAUGCCGCAA-AGCAUUUCCAGCUUAAUGAUUGCAUUCUUGC-----A----- .(((((...---.((((((((((((.((((..........(((....))).....))))..)))).))-)))))).)))))........((((....)))-----)----- ( -26.96) >DroSec_CAF1 1256 102 + 1 UGUUGGGAUCUCUAGAACU---UGC-UGCAUUUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA-AGCUUUUC--GCUUAAUGACGGCAUUUUCUUCG-C-AUUUUC ....(((.((....))..(---(((-((...........))))))...)))(((..((.(((((((((-(((.....--))))..)).))))))..))..))-)-...... ( -25.30) >DroSim_CAF1 10925 102 + 1 UGUUGGGAUCUCUAGAACU---UGC-UGCAUUUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA-AGCUUUUC--GCUUAAUGACGGCAUUUUCCUCG-C-AUUUUC (((.((((.......(((.---((.-.(((((((........))))))).)).)))...(((((((((-(((.....--))))..)).))))))..)))).)-)-)..... ( -26.80) >DroYak_CAF1 11663 103 + 1 UGUUGGGAUCUGUAGAACUU--UGC-UGCAUUUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA-AGCUUUCC--GCUUAAUGAUGGCAUUUUCUUCG-C-AUUUUC (((..(((...(.(((.(((--(((-.(((((..((....(((....)))...))..).)))).))))-)).))).)--(((.......)))....)))..)-)-)..... ( -24.90) >consensus UGUUGGGAUCUCUAGAACU___UGC_UGCAUUUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA_AGCUUUUC__GCUUAAUGACGGCAUUUUCCUCG_C_AUUUUC ....(((....................(((((((........)))))))..........((((((((..(((.......)))...)).))))))....))).......... (-16.04 = -15.85 + -0.19)


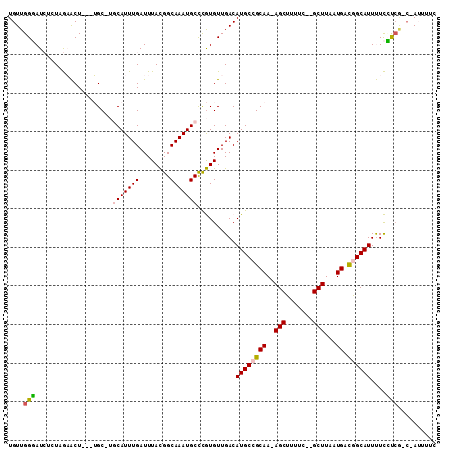
| Location | 14,086,023 – 14,086,125 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -14.60 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14086023 102 - 20766785 GAAAAU-G-CGAGGAAAAUGCCGUCAUUAAGC--GAAAAGCU-UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAAAUGCA-GCA---AGUUCUAGAGAUCCCAACA (((..(-(-(..((...(((((((....((((--.....)))-).))))))).)).......(((((((........))))))).-)))---..))).............. ( -28.20) >DroVir_CAF1 2089 103 - 1 -----UGGCUGGCAAGAAUGCAAUCAUUAAGCUGGAAAUGCUCUUGCGGCAUGUCAACAUAAGCAUUUGCCAUAAAUCAAAUGACGGCCAAAAGCAUUA---AUGCCAACA -----..(.(((((..(((((.........((.(((.....))).))((((((....)))..(((((((........)))))).).)))....))))).---.))))).). ( -27.50) >DroGri_CAF1 2320 97 - 1 -----U-----GCAAGAAUGCAAUCAUUAAGCUGGAAAUGCU-UUGCGGCAUGUCAACAUAAGCAUUUGCCAUAAAUCAAAUGACGGCCAAAAGCAUUA---AUGCCAACA -----(-----(((....))))........(.(((.((((((-((..((((((....)))..(((((((........)))))).).))).)))))))).---...))).). ( -23.20) >DroSec_CAF1 1256 102 - 1 GAAAAU-G-CGAAGAAAAUGCCGUCAUUAAGC--GAAAAGCU-UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAAAUGCA-GCA---AGUUCUAGAGAUCCCAACA (((..(-(-(((.....(((((((....((((--.....)))-).))))))).)).......(((((((........))))))).-)))---..))).............. ( -27.90) >DroSim_CAF1 10925 102 - 1 GAAAAU-G-CGAGGAAAAUGCCGUCAUUAAGC--GAAAAGCU-UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAAAUGCA-GCA---AGUUCUAGAGAUCCCAACA (((..(-(-(..((...(((((((....((((--.....)))-).))))))).)).......(((((((........))))))).-)))---..))).............. ( -28.20) >DroYak_CAF1 11663 103 - 1 GAAAAU-G-CGAAGAAAAUGCCAUCAUUAAGC--GGAAAGCU-UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAAAUGCA-GCA--AAGUUCUACAGAUCCCAACA ......-.-.(.((((..(((...((((..((--.....))(-((((((((((((......))))..)))))))))...))))..-)))--...)))).)........... ( -23.30) >consensus GAAAAU_G_CGAAGAAAAUGCCAUCAUUAAGC__GAAAAGCU_UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAAAUGCA_GCA___AGUUCUAGAGAUCCCAACA ............((((..(((...((((.(((.......))).((((((((((((......))))..))))))))....))))...))).....))))............. (-14.60 = -15.27 + 0.67)

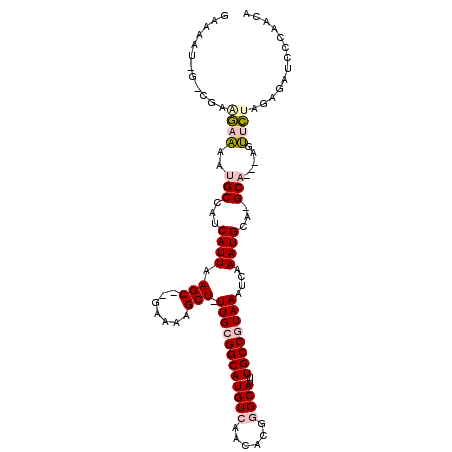
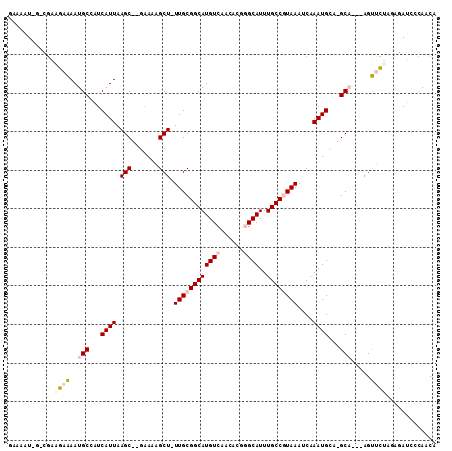
| Location | 14,086,050 – 14,086,145 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.46 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -13.31 |
| Energy contribution | -12.73 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
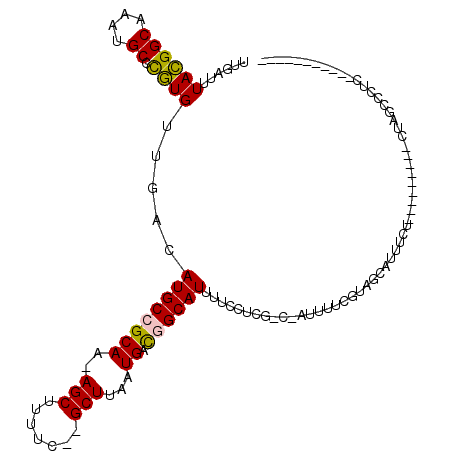
>2R_DroMel_CAF1 14086050 95 + 20766785 UUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA-AGCUUUUC--GCUUAAUGACGGCAUUUUCCUCG-C-AUUUUCGUAGCAUUUUU---------CUAGCCCUC----------- .....((((((.((((((....(..((.(((((((((-(((.....--))))..)).))))))))..)..)-)-)))))))))).......---------.........----------- ( -22.60) >DroVir_CAF1 2117 95 + 1 UUGAUUUAUGGCAAAUGCUUAUGUUGACAUGCCGCAAGAGCAUUUCCAGCUUAAUGAUUGCAUUCUUGCCAGCCA-------GCCAUUUCUAUUUGG-------GCUGU----------- ........(((((((((((((.((((..(((((....).))))...)))).))).....))))..))))))..((-------(((...........)-------)))).----------- ( -26.30) >DroGri_CAF1 2348 96 + 1 UUGAUUUAUGGCAAAUGCUUAUGUUGACAUGCCGCAA-AGCAUUUCCAGCUUAAUGAUUGCAUUCUUGC-----A-------ACCAUUUCUAUUUGGAUACCAGGCCAG----------- ........((((.(((((((.(((.(.....).))))-))))))(((((...((((.(((((....)))-----)-------).)))).....)))))......)))).----------- ( -24.90) >DroSec_CAF1 1283 95 + 1 UUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA-AGCUUUUC--GCUUAAUGACGGCAUUUUCUUCG-C-AUUUUCGUAGCAUUUUU---------CUAGCCCUC----------- .....((((((.((((((..((....))(((((((((-(((.....--))))..)).)))))).......)-)-)))))))))).......---------.........----------- ( -22.20) >DroSim_CAF1 10952 95 + 1 UUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA-AGCUUUUC--GCUUAAUGACGGCAUUUUCCUCG-C-AUUUUCGUAGCAUUUCU---------CUAGCCCUC----------- .....((((((.((((((....(..((.(((((((((-(((.....--))))..)).))))))))..)..)-)-)))))))))).......---------.........----------- ( -22.60) >DroYak_CAF1 11691 106 + 1 UUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA-AGCUUUCC--GCUUAAUGAUGGCAUUUUCUUCG-C-AUUUUCGUAGCAUUUUC---------CUCGCAUUUUCGUAGCAUUU .....((((((.((((((..(((..((.(((((((((-(((.....--))))..)).))))))..))..))-)-......(.((.......---------)))))))))))))))..... ( -22.60) >consensus UUGAUUUACGGCAAAUGCCCGUGUUGACAUGCCGCAA_AGCUUUUC__GCUUAAUGACGGCAUUUUCCUCG_C_AUUUUCGUAGCAUUUCU_________CUAGCCCUC___________ ......((((((....)).)))).....((((((((..(((.......)))...)).))))))......................................................... (-13.31 = -12.73 + -0.58)
| Location | 14,086,050 – 14,086,145 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.46 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -11.03 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14086050 95 - 20766785 -----------GAGGGCUAG---------AAAAAUGCUACGAAAAU-G-CGAGGAAAAUGCCGUCAUUAAGC--GAAAAGCU-UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAA -----------...(((...---------..(((((((.((.....-.-)).((...(((((((....((((--.....)))-).))))))).))......))))))))))......... ( -25.60) >DroVir_CAF1 2117 95 - 1 -----------ACAGC-------CCAAAUAGAAAUGGC-------UGGCUGGCAAGAAUGCAAUCAUUAAGCUGGAAAUGCUCUUGCGGCAUGUCAACAUAAGCAUUUGCCAUAAAUCAA -----------.((((-------(...........)))-------))..((((((..((((.........((((.((......)).))))(((....)))..))))))))))........ ( -24.80) >DroGri_CAF1 2348 96 - 1 -----------CUGGCCUGGUAUCCAAAUAGAAAUGGU-------U-----GCAAGAAUGCAAUCAUUAAGCUGGAAAUGCU-UUGCGGCAUGUCAACAUAAGCAUUUGCCAUAAAUCAA -----------.(((((..((.((......))((((((-------(-----(((....))))))))))..))..)(((((((-(((..(.....)..)).))))))))))))........ ( -25.30) >DroSec_CAF1 1283 95 - 1 -----------GAGGGCUAG---------AAAAAUGCUACGAAAAU-G-CGAAGAAAAUGCCGUCAUUAAGC--GAAAAGCU-UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAA -----------((.(((...---------.....(((.........-)-))........))).))....(((--.....)))-((((((((((((......))))..))))))))..... ( -25.59) >DroSim_CAF1 10952 95 - 1 -----------GAGGGCUAG---------AGAAAUGCUACGAAAAU-G-CGAGGAAAAUGCCGUCAUUAAGC--GAAAAGCU-UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAA -----------...(((...---------..(((((((.((.....-.-)).((...(((((((....((((--.....)))-).))))))).))......))))))))))......... ( -25.60) >DroYak_CAF1 11691 106 - 1 AAAUGCUACGAAAAUGCGAG---------GAAAAUGCUACGAAAAU-G-CGAAGAAAAUGCCAUCAUUAAGC--GGAAAGCU-UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAA ....(((.((..((((...(---------(......((.((.....-.-)).))......))..))))...)--)...)))(-((((((((((((......))))..))))))))).... ( -23.80) >consensus ___________GAGGGCUAG_________AAAAAUGCUACGAAAAU_G_CGAAGAAAAUGCCAUCAUUAAGC__GAAAAGCU_UUGCGGCAUGUCAACACGGGCAUUUGCCGUAAAUCAA .....................................................................(((.......))).((((((((((((......))))..))))))))..... (-11.03 = -11.70 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:09 2006