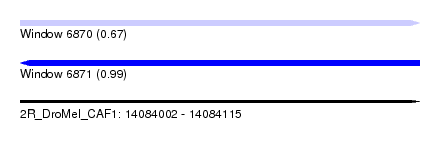
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,084,002 – 14,084,115 |
| Length | 113 |
| Max. P | 0.990099 |

| Location | 14,084,002 – 14,084,115 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.94 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -15.55 |
| Energy contribution | -16.33 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14084002 113 + 20766785 CCUGCUUAAGUA---ACCACUUUCAGCGGUUUGGCAAUAAAGCGAAUUUUUAUUGC----UAUUUGCAGGAGGAAAUCUAAAUUUCGUUUUCAAGAUUUUCUACUGAGUGGUCCUAAGUC ...(((((.(..---(((((((...((((..(((((((((((.....)))))))))----)).))))((.((((((((((((......)))..))))))))).)))))))))).))))). ( -31.50) >DroSim_CAF1 8985 113 + 1 CCUGCUUAAGUA---ACCACUUUCAGCGGUUUGGCAGUAAAGUGAAUUUUUAUUGC----UAUUUGUAGGAGAAAAUCUAAAUUUCGUUUUCCAGAUUUUCCCCUGAGCGAUCCUAAGUC .(((((.((((.---...))))..)))))..(((((((((((.....)))))))))----)).(((((((.(((((((((((......)))..)))))))).)))..))))......... ( -27.90) >DroYak_CAF1 9623 119 + 1 CCUUCUGCAGUAGUUACUACUUUCAGCGGUUAAGUAAUAAAGUUCGUUUUUAUUGCAUUUUAUUUGUCCGAGAAAAUGGAAUUUG-UAAUUUUAGAUUUUCCCUUAAGCGCUUUAGUUUC ........(((((...)))))...((((.(((((((((((((.....))))))))).............(((((..((((((...-..))))))..)))))..)))).))))........ ( -20.50) >consensus CCUGCUUAAGUA___ACCACUUUCAGCGGUUUGGCAAUAAAGUGAAUUUUUAUUGC____UAUUUGUAGGAGAAAAUCUAAAUUUCGUUUUCAAGAUUUUCCCCUGAGCGAUCCUAAGUC .(((((.((((.......))))..)))))....(((((((((.....)))))))))..........((((.(((((((((((......)))..)))))))).)))).............. (-15.55 = -16.33 + 0.79)

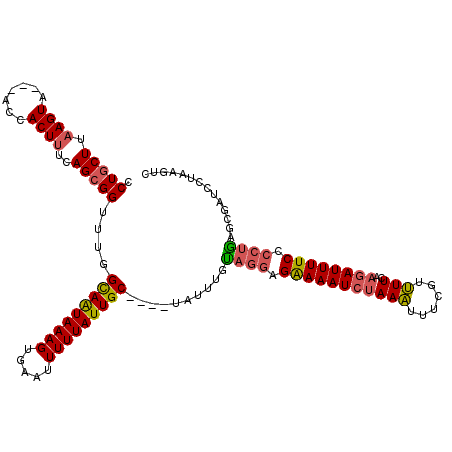
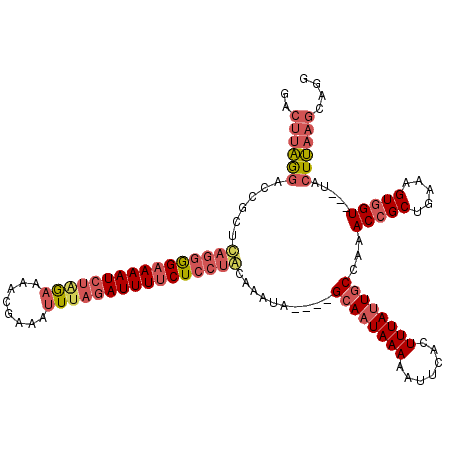
| Location | 14,084,002 – 14,084,115 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.94 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -15.77 |
| Energy contribution | -19.67 |
| Covariance contribution | 3.90 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14084002 113 - 20766785 GACUUAGGACCACUCAGUAGAAAAUCUUGAAAACGAAAUUUAGAUUUCCUCCUGCAAAUA----GCAAUAAAAAUUCGCUUUAUUGCCAAACCGCUGAAAGUGGU---UACUUAAGCAGG .(((.((.....)).)))((.((((((.((........)).)))))).))(((((.....----((((((((.......))))))))..((((((.....)))))---)......))))) ( -26.00) >DroSim_CAF1 8985 113 - 1 GACUUAGGAUCGCUCAGGGGAAAAUCUGGAAAACGAAAUUUAGAUUUUCUCCUACAAAUA----GCAAUAAAAAUUCACUUUACUGCCAAACCGCUGAAAGUGGU---UACUUAAGCAGG (.((((((.......(((((((((((((((........))))))))))))))).......----(((.((((.......)))).)))..((((((.....)))))---).)))))))... ( -32.00) >DroYak_CAF1 9623 119 - 1 GAAACUAAAGCGCUUAAGGGAAAAUCUAAAAUUA-CAAAUUCCAUUUUCUCGGACAAAUAAAAUGCAAUAAAAACGAACUUUAUUACUUAACCGCUGAAAGUAGUAACUACUGCAGAAGG ........((((.(((((................-.....(((........)))............((((((.......)))))).))))).))))....(((((....)))))...... ( -16.30) >consensus GACUUAGGACCGCUCAGGGGAAAAUCUAGAAAACGAAAUUUAGAUUUUCUCCUACAAAUA____GCAAUAAAAAUUCACUUUAUUGCCAAACCGCUGAAAGUGGU___UACUUAAGCAGG ..((((((......((((((((((((((((........))))))))))))))))..........((((((((.......))))))))...(((((.....))))).....)))))).... (-15.77 = -19.67 + 3.90)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:04 2006