
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,083,793 – 14,083,928 |
| Length | 135 |
| Max. P | 0.941946 |

| Location | 14,083,793 – 14,083,906 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
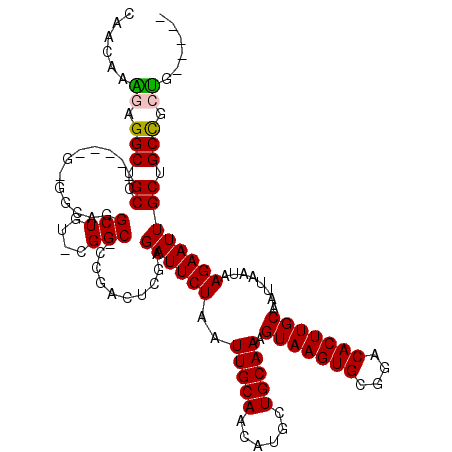
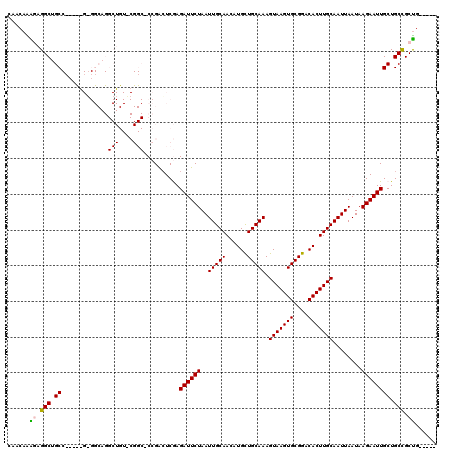
>2R_DroMel_CAF1 14083793 113 + 20766785 CAACAAAGAGGCUGCCCC-CGGGGGCAGGCUGUACGGC-CCGACUCGAGAUUCUAAUUGCAACAUGCUGCAAAGUAAGUGCGGACACUUGCAAUUAAUAAGAAUUGCUGCCGCUG----- ......(.((.((((((.-...)))))).)).).((((-.(((.((...((..(((((((((....(((((.......)))))....)))))))))))..)).)))..))))...----- ( -38.50) >DroPse_CAF1 800 96 + 1 CAACAGGCAGGCAGC------------------------CAGGCUCAAGAUUCUAAUUGCAACAUGUUGCAAAGUAAGUGCGGACACUUGCAAUUAAUAAGAAUUGCUGCUGCUGUGGCU .....(((.((((((------------------------..(((....((((((..((((((....)))))).(((((((....)))))))........)))))))))))))))...))) ( -33.10) >DroSim_CAF1 8778 111 + 1 CAACAAAGAGGCUGCC---UGGGGGCAGGCUGUUCGGC-CCGACUCGAGAUUCUAAUUGCAACAUGCUGCAAAGUAAGUGCGGACACUUGCAAUUAAUAAGAAUUGCUGCCGCUG----- ......((.(((.(((---(.(((.(.((((....)))-).).))).))(((((((((((((....(((((.......)))))....))))))))....))))).)).))).)).----- ( -35.10) >DroYak_CAF1 9405 115 + 1 CAACAAAGAGGCUGCCCCGCAGUGGCAGGCUGUACGGCUCCGAGUCGAGAUUCUAAUUGCAACAUGCUGCAAAGUAAGUGCGGACACUUGCAAUUAAUAAGAAUUGCUGCCGCUG----- .........((.....)).(((((((((......((((.....)))).((((((((((((((....(((((.......)))))....))))))))....)))))).)))))))))----- ( -37.10) >DroPer_CAF1 835 96 + 1 CAACAGGCAGGCAGC------------------------CAGGCUCAAGAUUCUAAUUGCAACAUGUUGCAAAGUAAGUGCGGACACUUGCAAUUAAUAAGAAUUGCUGCUGCCGUGGCU .....(((.((((((------------------------..(((....((((((..((((((....)))))).(((((((....)))))))........)))))))))))))))...))) ( -35.00) >consensus CAACAAAGAGGCUGCC_____G_GGCAGGCUGU_CGGC_CCGACUCGAGAUUCUAAUUGCAACAUGCUGCAAAGUAAGUGCGGACACUUGCAAUUAAUAAGAAUUGCUGCCGCUG_____ ......((.(((.((.............(((....)))..........((((((..(((((......))))).(((((((....)))))))........)))))))).))).))...... (-21.42 = -21.48 + 0.06)

| Location | 14,083,831 – 14,083,928 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.44 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
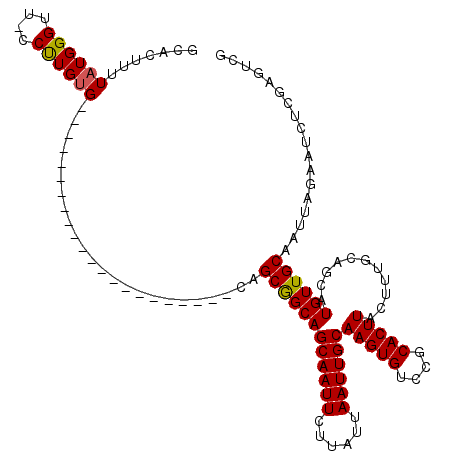
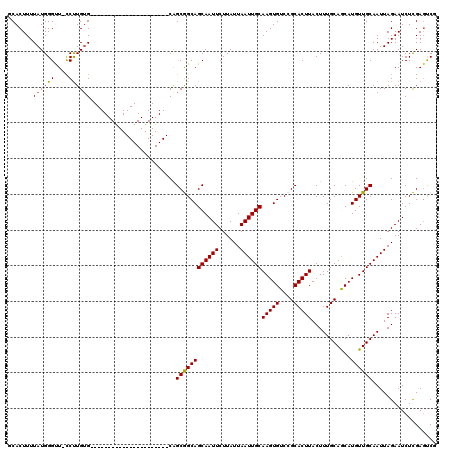
>2R_DroMel_CAF1 14083831 97 - 20766785 GCACUUUUAUGGGUU-CCUUGUG----------------------CAGCGGCAGCAAUUCUUAUUAAUUGCAAGUGUCCGCACUUACUUUGCAGCAUGUUGCAAUUAGAAUCUCGAGUCG (.((((....(((((-(....((----------------------(((((((.((((((......))))))(((((....)))))........)).)))))))....)))))).))))). ( -28.00) >DroPse_CAF1 816 119 - 1 UCACUUUUAUGGGCU-CCUUGGGCUCGGUGCUCUGGCAGCAGCCACAGCAGCAGCAAUUCUUAUUAAUUGCAAGUGUCCGCACUUACUUUGCAACAUGUUGCAAUUAGAAUCUUGAGCCU ..........(((((-(...((..(((.((((.((((....)))).)))).).((((((......))))))(((((....)))))...((((((....))))))...))..)).)))))) ( -40.60) >DroSim_CAF1 8814 97 - 1 GCACUUUUAUGGGUU-CCCUGUG----------------------CAGCGGCAGCAAUUCUUAUUAAUUGCAAGUGUCCGCACUUACUUUGCAGCAUGUUGCAAUUAGAAUCUCGAGUCG (.((((....(((((-(....((----------------------(((((((.((((((......))))))(((((....)))))........)).)))))))....)))))).))))). ( -28.00) >DroYak_CAF1 9445 98 - 1 GCACUUUUAUGGGUUUUCUUGUG----------------------CAGCGGCAGCAAUUCUUAUUAAUUGCAAGUGUCCGCACUUACUUUGCAGCAUGUUGCAAUUAGAAUCUCGACUCG ..........(((((((((..((----------------------(((((((.((((((......))))))(((((....)))))........)).)))))))...))))....))))). ( -28.10) >DroPer_CAF1 851 119 - 1 UCACUUUUAUGGGCU-CCUUGUGCUCGGUGUUCUGGCAGCAGCCACGGCAGCAGCAAUUCUUAUUAAUUGCAAGUGUCCGCACUUACUUUGCAACAUGUUGCAAUUAGAAUCUUGAGCCU ..........(((((-(....((((.(.(((..((((....))))..))).))))).......((((((((((((((..(((.......))).)))).))))))))))......)))))) ( -38.90) >consensus GCACUUUUAUGGGUU_CCUUGUG______________________CAGCGGCAGCAAUUCUUAUUAAUUGCAAGUGUCCGCACUUACUUUGCAGCAUGUUGCAAUUAGAAUCUCGAGUCG .......((((((....))))))........................((((((((((((......))))))(((((....)))))...........)))))).................. (-17.64 = -17.44 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:02 2006