
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,083,056 – 14,083,193 |
| Length | 137 |
| Max. P | 0.953236 |

| Location | 14,083,056 – 14,083,153 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -27.90 |
| Energy contribution | -27.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14083056 97 + 20766785 GUUCGGGCCAAGAUUGGCACAACGACCACCG---GCGUGGCAGCUG--------------------UUUUGUGGCAGCCAUAAUUGGCAUGUGGAAAACUGGCGGAUUGGCCAUAAAAAG .....((((((...((.(((..((.....))---..))).))((((--------------------((((.(.(((((((....)))).))).)))))).)))...))))))........ ( -32.90) >DroSim_CAF1 8034 97 + 1 CUUCGGGCCAAGAUUGGCACAACGACCACCG---GCGUGGCAGCUG--------------------UUUUGUGGCAGCCAUAAUUGGCAUGUGGAAAACUGGCGGAGUGGCCAUAAAAAG .....(((((....((.(((..((.....))---..))).))((((--------------------((((.(.(((((((....)))).))).)))))).)))....)))))........ ( -31.90) >DroYak_CAF1 8621 120 + 1 GUUCGGGCCAAGAUUGGCGCAACGACCACCGUGGUCGUGGCAGCUGUUUUCUGCUGUUUUAUGCUGUUUUGUGGCAGCCAUAAUUGGCAUGUGGAAAACUGGCGGCGUGGCCAUAAAAAG .....(((((....((.(((.((((((.....)))))).)).(((((((((..(((((((((((((((....))))).)))))..)))).)..)))))).)))).)))))))........ ( -46.80) >consensus GUUCGGGCCAAGAUUGGCACAACGACCACCG___GCGUGGCAGCUG____________________UUUUGUGGCAGCCAUAAUUGGCAUGUGGAAAACUGGCGGAGUGGCCAUAAAAAG ..(((.((((....))))....)))((((.......))))..........................(((((((((.((((....)))).(((.(.....).))).....))))))))).. (-27.90 = -27.90 + 0.00)

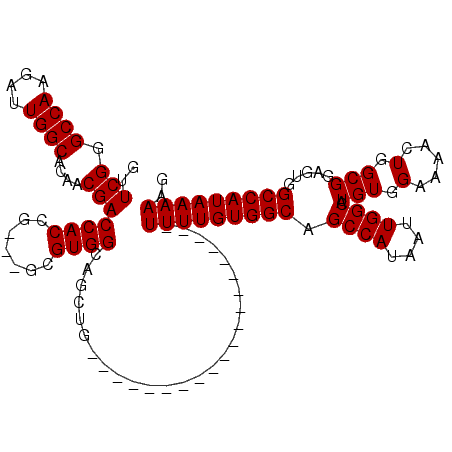
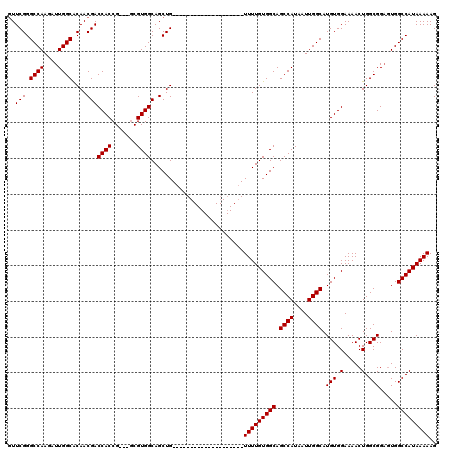
| Location | 14,083,056 – 14,083,153 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -24.90 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14083056 97 - 20766785 CUUUUUAUGGCCAAUCCGCCAGUUUUCCACAUGCCAAUUAUGGCUGCCACAAAA--------------------CAGCUGCCACGC---CGGUGGUCGUUGUGCCAAUCUUGGCCCGAAC .......((((......))))...........(((((...((((.........(--------------------((((.(((((..---..))))).)))))))))...)))))...... ( -27.90) >DroSim_CAF1 8034 97 - 1 CUUUUUAUGGCCACUCCGCCAGUUUUCCACAUGCCAAUUAUGGCUGCCACAAAA--------------------CAGCUGCCACGC---CGGUGGUCGUUGUGCCAAUCUUGGCCCGAAG .......((((......))))...........(((((...((((.........(--------------------((((.(((((..---..))))).)))))))))...)))))...... ( -27.90) >DroYak_CAF1 8621 120 - 1 CUUUUUAUGGCCACGCCGCCAGUUUUCCACAUGCCAAUUAUGGCUGCCACAAAACAGCAUAAAACAGCAGAAAACAGCUGCCACGACCACGGUGGUCGUUGCGCCAAUCUUGGCCCGAAC ........(((...)))((((((((((....(((...(((((.(((........))))))))....))))))))).((.((.((((((.....)))))).))))......))))...... ( -34.40) >consensus CUUUUUAUGGCCACUCCGCCAGUUUUCCACAUGCCAAUUAUGGCUGCCACAAAA____________________CAGCUGCCACGC___CGGUGGUCGUUGUGCCAAUCUUGGCCCGAAC .......((((......))))...........(((((...((((((...)).......................((((.(((((.......))))).)))).))))...)))))...... (-24.90 = -24.90 + 0.00)

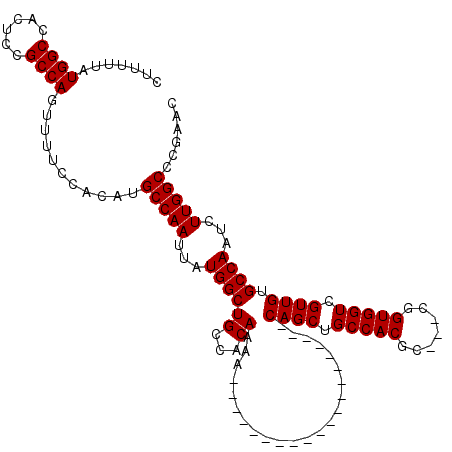
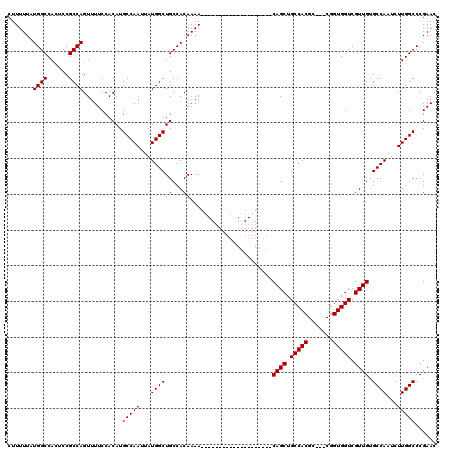
| Location | 14,083,093 – 14,083,193 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.47 |
| Mean single sequence MFE | -36.69 |
| Consensus MFE | -30.25 |
| Energy contribution | -30.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14083093 100 + 20766785 CAGCUG--------------------UUUUGUGGCAGCCAUAAUUGGCAUGUGGAAAACUGGCGGAUUGGCCAUAAAAAGCAGGUCAGGCCACUGCUAAGACAAUCAAAUAAUGGCCACU ..((((--------------------((((.(.(((((((....)))).))).)))))).)))....(((((((....(((((.........)))))..(.....).....))))))).. ( -31.00) >DroSim_CAF1 8071 100 + 1 CAGCUG--------------------UUUUGUGGCAGCCAUAAUUGGCAUGUGGAAAACUGGCGGAGUGGCCAUAAAAAGCAGGUCAGGCCACUGCUAAGACAAUCAAAUAAUGGCCACU ..((((--------------------((((.(.(((((((....)))).))).)))))).)))..(((((((((....(((((.........)))))..(.....).....))))))))) ( -35.00) >DroYak_CAF1 8661 120 + 1 CAGCUGUUUUCUGCUGUUUUAUGCUGUUUUGUGGCAGCCAUAAUUGGCAUGUGGAAAACUGGCGGCGUGGCCAUAAAAAGCAGGUCAGGCCACUGCCAAGACAAUCAAAUAAUGGCCACU ..(((((((((..(((((((((((((((....))))).)))))..)))).)..)))))).)))...((((((((.........(((.(((....)))..))).........)))))))). ( -44.07) >consensus CAGCUG____________________UUUUGUGGCAGCCAUAAUUGGCAUGUGGAAAACUGGCGGAGUGGCCAUAAAAAGCAGGUCAGGCCACUGCUAAGACAAUCAAAUAAUGGCCACU ..............................(((((.((((....))))...........((((((..(((((...............)))))))))))................))))). (-30.25 = -30.03 + -0.22)

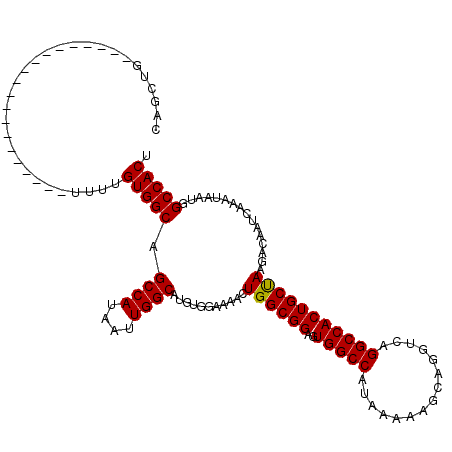
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:59 2006