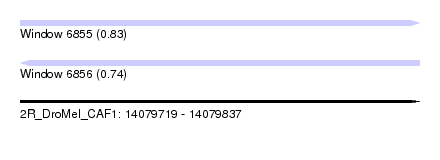
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,079,719 – 14,079,837 |
| Length | 118 |
| Max. P | 0.834892 |

| Location | 14,079,719 – 14,079,837 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.64 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -25.76 |
| Energy contribution | -25.10 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
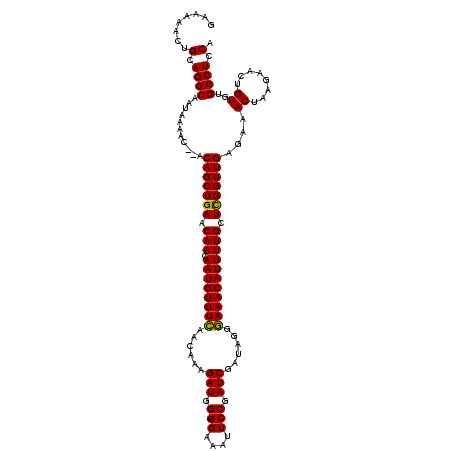
>2R_DroMel_CAF1 14079719 118 + 20766785 GAAGAACUGCAGGCAAUAAAAC--ACAGCUGAGCAACAAUGUUUCAACAAAGAUGGGGGAAAUUCCGAUCGAUAGGGAAACAUUUUGCUCGGUUGAGAAACUAAGAACUGUGUGCCUUCA ((((.((.((((..........--.(((((((((((.((((((((......(((.(((.....))).)))......)))))))))))))))))))............))))))..)))). ( -39.80) >DroSim_CAF1 4270 117 + 1 GAAAAACUGCAGGCAAUAAAAC--ACAGCUGAACAACAAUGUUUCAACAAAGAUGG-GGAAAUUCCGAUCGAUAGGGAAACAUUUUGCUCGGUUGAGAAACUAAGAACUGUGUGCCUGCA ........(((((((.......--.(((((((.(((.((((((((......(((.(-((....))).)))......))))))))))).)))))))....((........)).))))))). ( -38.80) >DroYak_CAF1 4942 119 + 1 GAAAAACUGCAGGCGAUAAAACACACAGCUGAACAACAAUGUUUUCACAGCGAUGG-GGAAAUUCCGAUCGAUAGGAAAACAUUUUGCUUGGUUGAGAAACUUCGAAGUGUAGGCCUCCA ........(.((((......((((.((((..(.(((.(((((((((....((((.(-((....))).))))....)))))))))))).)..)))).((....))...))))..)))).). ( -34.20) >consensus GAAAAACUGCAGGCAAUAAAAC__ACAGCUGAACAACAAUGUUUCAACAAAGAUGG_GGAAAUUCCGAUCGAUAGGGAAACAUUUUGCUCGGUUGAGAAACUAAGAACUGUGUGCCUCCA ........(.((((...........(((((((.(((.((((((((......(((.(.((....))).)))......))))))))))).)))))))....((........))..)))).). (-25.76 = -25.10 + -0.66)

| Location | 14,079,719 – 14,079,837 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.64 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.83 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
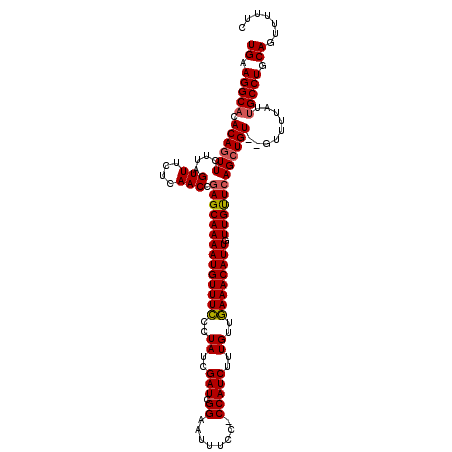
>2R_DroMel_CAF1 14079719 118 - 20766785 UGAAGGCACACAGUUCUUAGUUUCUCAACCGAGCAAAAUGUUUCCCUAUCGAUCGGAAUUUCCCCCAUCUUUGUUGAAACAUUGUUGCUCAGCUGU--GUUUUAUUGCCUGCAGUUCUUC ((.((((((((((((....(((....))).((((((((((((((..((..(((.((.......)).)))..))..)))))))).))))))))))))--))......)))).))....... ( -32.90) >DroSim_CAF1 4270 117 - 1 UGCAGGCACACAGUUCUUAGUUUCUCAACCGAGCAAAAUGUUUCCCUAUCGAUCGGAAUUUCC-CCAUCUUUGUUGAAACAUUGUUGUUCAGCUGU--GUUUUAUUGCCUGCAGUUUUUC (((((((((((((((....(((....))).((((((((((((((..((..(((.((......)-).)))..))..)))))))).))))))))))))--))......)))))))....... ( -37.60) >DroYak_CAF1 4942 119 - 1 UGGAGGCCUACACUUCGAAGUUUCUCAACCAAGCAAAAUGUUUUCCUAUCGAUCGGAAUUUCC-CCAUCGCUGUGAAAACAUUGUUGUUCAGCUGUGUGUUUUAUCGCCUGCAGUUUUUC ((.((((..((((.....((((...((((.......(((((((((.((.((((.((......)-).)))).)).)))))))))))))...))))..))))......)))).))....... ( -29.51) >consensus UGAAGGCACACAGUUCUUAGUUUCUCAACCGAGCAAAAUGUUUCCCUAUCGAUCGGAAUUUCC_CCAUCUUUGUUGAAACAUUGUUGUUCAGCUGU__GUUUUAUUGCCUGCAGUUUUUC ((.(((((.((((((....(((....))).((((((((((((((..((..(((.((........)))))..))..)))))))).)))))))))))).........))))).))....... (-22.50 = -22.83 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:49 2006