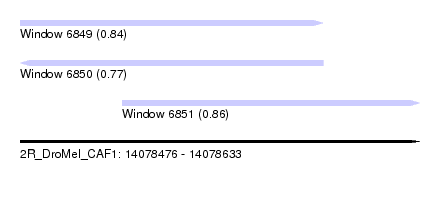
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,078,476 – 14,078,633 |
| Length | 157 |
| Max. P | 0.863282 |

| Location | 14,078,476 – 14,078,595 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -42.73 |
| Consensus MFE | -39.25 |
| Energy contribution | -39.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
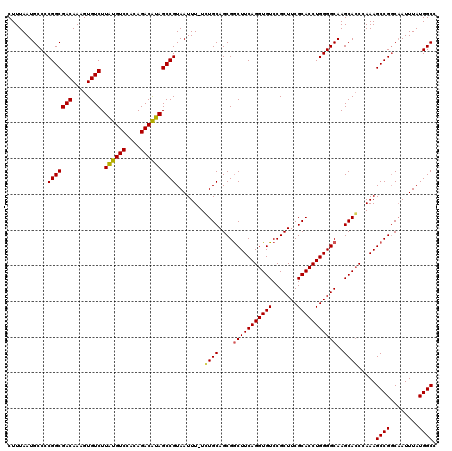
>2R_DroMel_CAF1 14078476 119 + 20766785 CUUUAAUGCCCCGGCGACAAAGUGUCUUAUGUCCACAGACAUAGCCGUAAUUU-UCUGCAGCGGCUUCAGGUGUCCGCUUCGCACCUGGGGCAAGCAGCAAAAGCCGGCAAUUUAUGGCC ......((((.(((((((.....))).((((((....))))))))))...(((-(((((....(((((((((((.......)))))))))))..)))).))))...)))).......... ( -44.10) >DroSim_CAF1 3027 119 + 1 CUUUAAUGCCCCGGCGACAAAGUGUCUUAUGUCCACAGACAUAGCCGUAAUUU-UCUGCAGCGACUUCAGGUGUCCGCUUCGCACCUGGGGCAAGCACCCAAAGCCGCCCAUUUAUGGCC .......(((.(((((((.....))).((((((....))))))))))......-......(((.(((..(((((..((((((....))))))..)))))..))).)))........))). ( -37.90) >DroYak_CAF1 3667 120 + 1 CUUUAAUGCCCCGGCGACAAAGUGUCUUGUGUCCACAGACGCAGCCGUAAUUUUUGUGCAGCGGCUUCAGGUGUCCGCUUCGCACCUGGGGCAAGCACCCAAAGCCGGCAAUUUAUGGCG .......(((.(((((((.....))).((((((....)))))))))).........(((..((((((..(((((..((((((....))))))..)))))..)))))))))......))). ( -46.20) >consensus CUUUAAUGCCCCGGCGACAAAGUGUCUUAUGUCCACAGACAUAGCCGUAAUUU_UCUGCAGCGGCUUCAGGUGUCCGCUUCGCACCUGGGGCAAGCACCCAAAGCCGGCAAUUUAUGGCC ...........(((((((.....))).((((((....))))))))))........((((....(((((((((((.......)))))))))))..)))).....((((........)))). (-39.25 = -39.03 + -0.22)

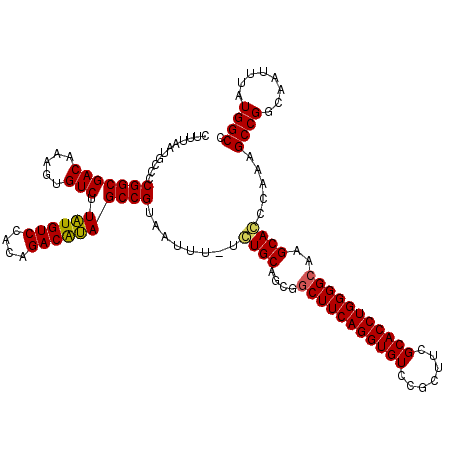
| Location | 14,078,476 – 14,078,595 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -40.53 |
| Energy contribution | -40.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14078476 119 - 20766785 GGCCAUAAAUUGCCGGCUUUUGCUGCUUGCCCCAGGUGCGAAGCGGACACCUGAAGCCGCUGCAGA-AAAUUACGGCUAUGUCUGUGGACAUAAGACACUUUGUCGCCGGGGCAUUAAAG .(((.....((((((((((((((((((((((....).)).)))))).))...)))))))..)))).-......((((((((((....)))))).(((.....))))))).)))....... ( -44.90) >DroSim_CAF1 3027 119 - 1 GGCCAUAAAUGGGCGGCUUUGGGUGCUUGCCCCAGGUGCGAAGCGGACACCUGAAGUCGCUGCAGA-AAAUUACGGCUAUGUCUGUGGACAUAAGACACUUUGUCGCCGGGGCAUUAAAG .(((.......((((((((..((((..(((..(......)..)))..))))..)))))))).....-......((((((((((....)))))).(((.....))))))).)))....... ( -47.80) >DroYak_CAF1 3667 120 - 1 CGCCAUAAAUUGCCGGCUUUGGGUGCUUGCCCCAGGUGCGAAGCGGACACCUGAAGCCGCUGCACAAAAAUUACGGCUGCGUCUGUGGACACAAGACACUUUGUCGCCGGGGCAUUAAAG .(((......(((((((((..((((..(((..(......)..)))..))))..))))))..))).........((((((.(((....))).)).(((.....))))))).)))....... ( -45.60) >consensus GGCCAUAAAUUGCCGGCUUUGGGUGCUUGCCCCAGGUGCGAAGCGGACACCUGAAGCCGCUGCAGA_AAAUUACGGCUAUGUCUGUGGACAUAAGACACUUUGUCGCCGGGGCAUUAAAG .(((.......(.((((((..((((..(((..(......)..)))..))))..))))))).............((((((((((....)))))).(((.....))))))).)))....... (-40.53 = -40.53 + 0.00)

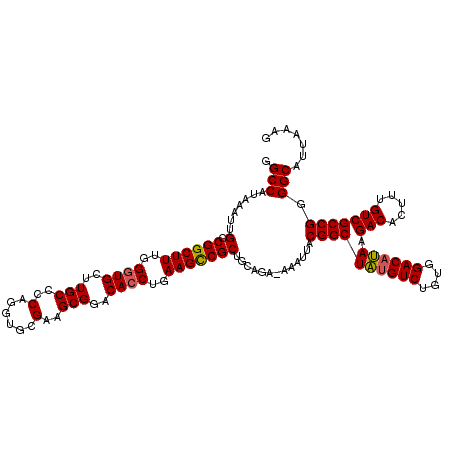
| Location | 14,078,516 – 14,078,633 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -42.97 |
| Consensus MFE | -36.66 |
| Energy contribution | -37.11 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14078516 117 + 20766785 AUAGCCGUAAUUU-UCUGCAGCGGCUUCAGGUGUCCGCUUCGCACCUGGGGCAAGCAGCAAAAGCCGGCAAUUUAUGGCCGGCCUGACACAAUUUCAUAAGGCCCC-AAAAGGCUC-CAU ...((((...(((-(((((....(((((((((((.......)))))))))))..)))).))))..)))).......(((((((((..............)))))..-....)))).-... ( -42.94) >DroSim_CAF1 3067 116 + 1 AUAGCCGUAAUUU-UCUGCAGCGACUUCAGGUGUCCGCUUCGCACCUGGGGCAAGCACCCAAAGCCGCCCAUUUAUGGCCGGCCUGACACAAUUUCAUAAGGCCCC-A-AAGGCUC-CAU ..(((((((....-..))).(((.(((..(((((..((((((....))))))..)))))..))).))).......(((..(((((..............)))))))-)-..)))).-... ( -35.54) >DroYak_CAF1 3707 120 + 1 GCAGCCGUAAUUUUUGUGCAGCGGCUUCAGGUGUCCGCUUCGCACCUGGGGCAAGCACCCAAAGCCGGCAAUUUAUGGCGGGCCUGACACAAUUUCAUAAGGCCCCCAAAAGGCUCGCAU ((((((..........(((..((((((..(((((..((((((....))))))..)))))..))))))))).....(((.((((((..............)))))))))...)))).)).. ( -50.44) >consensus AUAGCCGUAAUUU_UCUGCAGCGGCUUCAGGUGUCCGCUUCGCACCUGGGGCAAGCACCCAAAGCCGGCAAUUUAUGGCCGGCCUGACACAAUUUCAUAAGGCCCC_AAAAGGCUC_CAU ..((((.........((((....(((((((((((.......)))))))))))..)))).....((((........)))).(((((..............))))).......))))..... (-36.66 = -37.11 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:45 2006