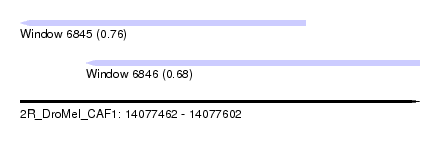
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,077,462 – 14,077,602 |
| Length | 140 |
| Max. P | 0.758317 |

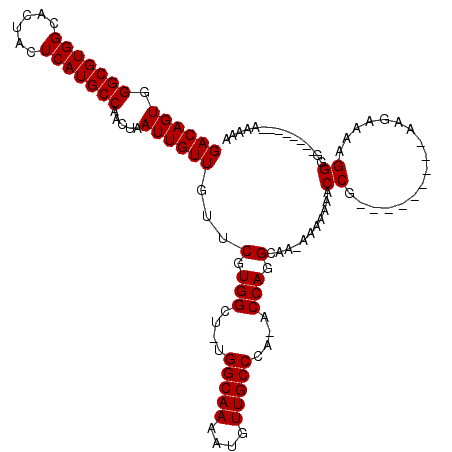
| Location | 14,077,462 – 14,077,562 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.71 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -22.43 |
| Energy contribution | -22.43 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14077462 100 - 20766785 GACAGUGGGCGUGGCACUACUCAUGCCAACUAAUUGUUGUUCGUGGCU-UGGCAAAAUGUUGCCCA-ACCAGGCAA-AAAAAAACCG---------AAGAAAAGGGG--------AAAAA ((((((.(((((((......))))))).....))))))...(.(((..-.(((((....)))))..-.))).)...-.......((.---------.......))..--------..... ( -24.20) >DroSim_CAF1 2000 100 - 1 GACAGUGGGCGUGGCACUACUCAUGCCAACUAAUUGUUGUUCGUGGCU-UGGCAAAAUGUUGCCCA-ACCAGGCAACAAAAAAACCG---------AAGAAAAGGGG--------AA-AA ...(((.(((((((......))))))).)))..((((((((..(((..-.(((((....)))))..-.))))))))))).....((.---------.......))..--------..-.. ( -29.10) >DroYak_CAF1 2255 115 - 1 GACAGUGGGCGUGGCACUACUCAUGCCAACUAAUUGUUGUUCGUGGCUUUGGCAAAAUGUUGCCCAAACCAGGCAA-A----AACCGUAGAAGAAAAAAAAAAGGGGCGACAAAGAAAAA ...(((.(((((((......))))))).)))..(((((((((.(((.((.(((((....))))).)).)))((...-.----..))..................)))))))))....... ( -31.60) >consensus GACAGUGGGCGUGGCACUACUCAUGCCAACUAAUUGUUGUUCGUGGCU_UGGCAAAAUGUUGCCCA_ACCAGGCAA_AAAAAAACCG_________AAGAAAAGGGG________AAAAA ((((((.(((((((......))))))).....))))))...(.(((....(((((....)))))....))).)...........((.................))............... (-22.43 = -22.43 + -0.00)

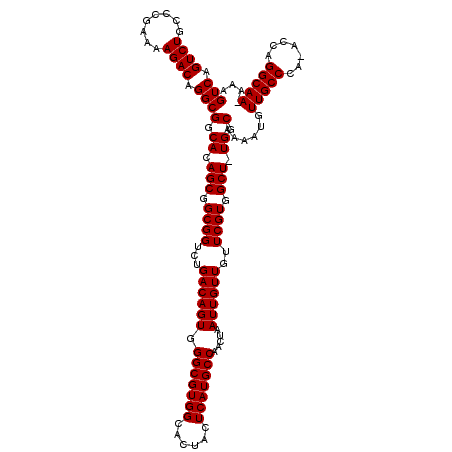
| Location | 14,077,485 – 14,077,602 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -43.63 |
| Consensus MFE | -39.40 |
| Energy contribution | -39.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14077485 117 - 20766785 GUCAGUCUGCCCGAAAAGACAGGCGGCACAGCGGCGGUCUGACAGUGGGCGUGGCACUACUCAUGCCAACUAAUUGUUGUUCGUGGCU-UGGCAAAAUGUUGCCCA-ACCAGGCAA-AAA ....(((.(((((((.((((..((.((...)).)).))))((((((.(((((((......))))))).....)))))).)))).))).-.)))......(((((..-....)))))-... ( -40.60) >DroSim_CAF1 2022 118 - 1 GUCAGUCUGCCCGAAAAGACAGGCGGCACAGCGGCGGUCUGACAGUGGGCGUGGCACUACUCAUGCCAACUAAUUGUUGUUCGUGGCU-UGGCAAAAUGUUGCCCA-ACCAGGCAACAAA (((((.((((((.....)....((......))))))).)))))(((.(((((((......))))))).)))..((((((((..(((..-.(((((....)))))..-.))))))))))). ( -45.20) >DroYak_CAF1 2293 117 - 1 GUCAGUCUGCCCGAAAAGACUGGCGGCACAGCGGCGGUCUGACAGUGGGCGUGGCACUACUCAUGCCAACUAAUUGUUGUUCGUGGCUUUGGCAAAAUGUUGCCCAAACCAGGCAA-A-- ((((((((........))))))))(.((.(((.((((...((((((.(((((((......))))))).....))))))..)))).))).)).)......(((((.......)))))-.-- ( -45.10) >consensus GUCAGUCUGCCCGAAAAGACAGGCGGCACAGCGGCGGUCUGACAGUGGGCGUGGCACUACUCAUGCCAACUAAUUGUUGUUCGUGGCU_UGGCAAAAUGUUGCCCA_ACCAGGCAA_AAA (((.((((........)))).)))(.((.(((.((((...((((((.(((((((......))))))).....))))))..)))).))).)).)......(((((.......))))).... (-39.40 = -39.40 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:40 2006