
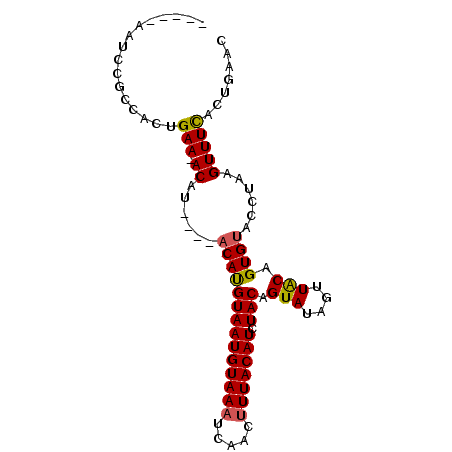
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,038,768 – 14,038,858 |
| Length | 90 |
| Max. P | 0.706047 |

| Location | 14,038,768 – 14,038,858 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -15.76 |
| Consensus MFE | -8.19 |
| Energy contribution | -8.12 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14038768 90 + 20766785 --GAAAAUCCGCCACUGAAUACAUGAACACAUGUAAUGUAAAUAAACUUUACAUCUACAGUAUAGUUACAGUGUACUUAAGUUUCUCUGAAC --((((......(((((..((((((....)))))).........((((.(((.......))).)))).)))))........))))....... ( -15.64) >DroSec_CAF1 9065 82 + 1 -----AAUCCGCCACUGAA-ACAU----ACAUGUAAUGUAAAUCAACUUUACAUCUACCGUAUAGUUACAGUGUACCUAAGUUUCACUGAAC -----.......(((((.(-((((----((..((((((((((.....))))))).))).)))).))).)))))................... ( -15.90) >DroSim_CAF1 4713 76 + 1 -----AAUCCGCCACUGAA-ACAU----ACAUGUAAUGUAAAUCAACUUUACAUCUACAGUAUAGUUACAGUGUACCUAAGUUUUA------ -----.......(((((.(-((((----((.(((((((((((.....))))))).)))))))).))).))))).............------ ( -18.50) >DroEre_CAF1 6007 87 + 1 UAAAAAAUCCGCCACCGAA-ACAU----ACACGUAAUGUAAAUGAAUCUUACAUCUACAGUAUAGUUGCAGUGCAUCUGAGUUUCACUGAAC ................(((-((..----....(((((((((.......)))))).)))....(((.(((...))).))).)))))....... ( -13.00) >consensus _____AAUCCGCCACUGAA_ACAU____ACAUGUAAUGUAAAUCAACUUUACAUCUACAGUAUAGUUACAGUGUACCUAAGUUUCACUGAAC ................(((.((......((((((((((((((.....))))))).))).(((....))).))))......)))))....... ( -8.19 = -8.12 + -0.06)

| Location | 14,038,768 – 14,038,858 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -12.85 |
| Energy contribution | -12.60 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14038768 90 - 20766785 GUUCAGAGAAACUUAAGUACACUGUAACUAUACUGUAGAUGUAAAGUUUAUUUACAUUACAUGUGUUCAUGUAUUCAGUGGCGGAUUUUC-- .....(((((.((...((.(((((..((.(((((((.((((((((.....))))))))))).))))....))...))))))))).)))))-- ( -18.40) >DroSec_CAF1 9065 82 - 1 GUUCAGUGAAACUUAGGUACACUGUAACUAUACGGUAGAUGUAAAGUUGAUUUACAUUACAUGU----AUGU-UUCAGUGGCGGAUU----- ((.((.((((((.((.((..((((((....)))))).((((((((.....))))))))..)).)----).))-)))).)))).....----- ( -21.00) >DroSim_CAF1 4713 76 - 1 ------UAAAACUUAGGUACACUGUAACUAUACUGUAGAUGUAAAGUUGAUUUACAUUACAUGU----AUGU-UUCAGUGGCGGAUU----- ------..........((.(((((.(((.(((((((.((((((((.....))))))))))).))----))))-).))))))).....----- ( -20.40) >DroEre_CAF1 6007 87 - 1 GUUCAGUGAAACUCAGAUGCACUGCAACUAUACUGUAGAUGUAAGAUUCAUUUACAUUACGUGU----AUGU-UUCGGUGGCGGAUUUUUUA ((.((.((((((....((((((((((.......))))((((((((.....))))))))..))))----))))-)))).)))).......... ( -18.70) >consensus GUUCAGUGAAACUUAGGUACACUGUAACUAUACUGUAGAUGUAAAGUUGAUUUACAUUACAUGU____AUGU_UUCAGUGGCGGAUU_____ ...................(((((..((...(((((.((((((((.....))))))))))).))......))...)))))............ (-12.85 = -12.60 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:23 2006