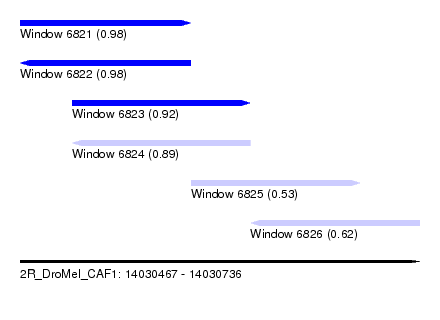
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,030,467 – 14,030,736 |
| Length | 269 |
| Max. P | 0.980419 |

| Location | 14,030,467 – 14,030,582 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -35.19 |
| Energy contribution | -35.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14030467 115 + 20766785 AUAAA-----AUGCAUUGGGGCUGGCAAUGGCAUUGGCCGAGGGGAUGUUUUGGGCCAAAAAAGCUCCAACCCACCCCAACGACCUCAACAGGCCAACAGUUGAUGCUGUGCCAAAAAAA .....-----.....((((.((.((((.((((.((((((..((((.((..((((((.......)).))))..))))))...(....)....))))))..)))).)))))).))))..... ( -38.50) >DroSim_CAF1 60549 115 + 1 AUAAA-----AUGCAUUGGGGCUGGCAAUGGCAUUGGCCGAGGGGAUGUUUUGGGCCAAAAAAGCUCCAACCCUCCCCAUCGACCUCAACAGGCCAACAGUUGAUGCUGUGCCAAAAAAA .....-----.....((((.((.((((.((((.(((((((((((((.(..((((((.......)).))))..)))))).))(....)....))))))..)))).)))))).))))..... ( -39.50) >DroEre_CAF1 63780 114 + 1 AUAAA----AAAGCAUUGGGGCUGGCAAUGGCAUUGGCCGAGGGGAUGUUUUGGGCCAA-AAAGCUC-UACCAUCCCCAUCGACCUCAACAGGCCAGCAGUUGAUGCUGUGCCAAAAAAA .....----..(((((..(.((((((..((.....((.((((((((((....((((...-...))))-...))))))).))).))....)).))))))..)..)))))............ ( -45.40) >DroYak_CAF1 62416 118 + 1 AUAAAGAAGAAAGCAUUGGGGCUGGCAAUGGCAUUGGCCGAGGGGAUGUUUUGGGCCAAAAAAGCUC-AACCCUCCCCAUCGACCUCAACAGGCCAACAGUUGAUGCUGUGCCAAA-AAU ........(..(((......)))..)..((((..((((((((((((.(..((((((.......))))-))..)))))).))(....)....)))))(((((....)))))))))..-... ( -41.00) >consensus AUAAA_____AAGCAUUGGGGCUGGCAAUGGCAUUGGCCGAGGGGAUGUUUUGGGCCAAAAAAGCUC_AACCCUCCCCAUCGACCUCAACAGGCCAACAGUUGAUGCUGUGCCAAAAAAA ...............((((.((.((((.((((.((((((..(((((.(.((.((((.......)))).)).).)))))...(....)....))))))..)))).)))))).))))..... (-35.19 = -35.50 + 0.31)

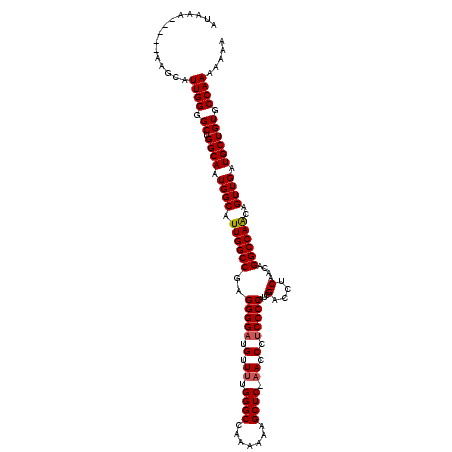
| Location | 14,030,467 – 14,030,582 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -36.92 |
| Energy contribution | -38.17 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14030467 115 - 20766785 UUUUUUUGGCACAGCAUCAACUGUUGGCCUGUUGAGGUCGUUGGGGUGGGUUGGAGCUUUUUUGGCCCAAAACAUCCCCUCGGCCAAUGCCAUUGCCAGCCCCAAUGCAU-----UUUAU ......(((((..((((((((.........)))))(((((..((((((..((((.(((.....)))))))..))))))..)))))..)))...)))))((......))..-----..... ( -44.80) >DroSim_CAF1 60549 115 - 1 UUUUUUUGGCACAGCAUCAACUGUUGGCCUGUUGAGGUCGAUGGGGAGGGUUGGAGCUUUUUUGGCCCAAAACAUCCCCUCGGCCAAUGCCAUUGCCAGCCCCAAUGCAU-----UUUAU ......(((((..((((((((.........)))))((((((.((((((..((((.(((.....)))))))..).)))))))))))..)))...)))))((......))..-----..... ( -44.70) >DroEre_CAF1 63780 114 - 1 UUUUUUUGGCACAGCAUCAACUGCUGGCCUGUUGAGGUCGAUGGGGAUGGUA-GAGCUUU-UUGGCCCAAAACAUCCCCUCGGCCAAUGCCAUUGCCAGCCCCAAUGCUUU----UUUAU ............(((((.....((((((..((...((((((.(((((((...-(.(((..-..))).)....)))))))))))))...))....))))))....)))))..----..... ( -41.50) >DroYak_CAF1 62416 118 - 1 AUU-UUUGGCACAGCAUCAACUGUUGGCCUGUUGAGGUCGAUGGGGAGGGUU-GAGCUUUUUUGGCCCAAAACAUCCCCUCGGCCAAUGCCAUUGCCAGCCCCAAUGCUUUCUUCUUUAU ...-.((((((..((((((((.........)))))((((((.((((((..((-(.(((.....))).)))..).)))))))))))..)))...))))))..................... ( -41.20) >consensus UUUUUUUGGCACAGCAUCAACUGUUGGCCUGUUGAGGUCGAUGGGGAGGGUU_GAGCUUUUUUGGCCCAAAACAUCCCCUCGGCCAAUGCCAUUGCCAGCCCCAAUGCAU_____UUUAU ......(((((..((((((((.........)))))((((((.(((((...((((.(((.....)))))))....)))))))))))..)))...)))))((......))............ (-36.92 = -38.17 + 1.25)


| Location | 14,030,502 – 14,030,622 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -34.41 |
| Energy contribution | -34.73 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14030502 120 + 20766785 AGGGGAUGUUUUGGGCCAAAAAAGCUCCAACCCACCCCAACGACCUCAACAGGCCAACAGUUGAUGCUGUGCCAAAAAAAUAGUGGGAAGCACAAGGAAUUGAACUUUGUGCCCCGUCUG .((((.((..((((((.......)).))))..))))))...(((.......(((..(((((....))))))))...........(((..((((((((.......)))))))))))))).. ( -42.10) >DroSim_CAF1 60584 119 + 1 AGGGGAUGUUUUGGGCCAAAAAAGCUCCAACCCUCCCCAUCGACCUCAACAGGCCAACAGUUGAUGCUGUGCCAAAAAAAUAGUGGGAAGCACAAGGAAUUGAACUUUGUGC-CCGUCUG .(((((.(..((((((.......)).))))..))))))...(((.......(((..(((((....))))))))............((..((((((((.......))))))))-))))).. ( -39.00) >DroEre_CAF1 63816 117 + 1 AGGGGAUGUUUUGGGCCAA-AAAGCUC-UACCAUCCCCAUCGACCUCAACAGGCCAGCAGUUGAUGCUGUGCCAAAAAAAUAGUGGGAAGCACAAGGAAUUGAACUUUGUGC-CCGCCUG .(((((((....((((...-...))))-...))))))).............((((((((.....))))).))).........((((...((((((((.......))))))))-))))... ( -44.90) >DroYak_CAF1 62456 117 + 1 AGGGGAUGUUUUGGGCCAAAAAAGCUC-AACCCUCCCCAUCGACCUCAACAGGCCAACAGUUGAUGCUGUGCCAAA-AAUUAGUGGGUAGCACAAGGAAUUGAACUUUGUGC-CCGUCUG .(((((.(..((((((.......))))-))..))))))...(((.......(((..(((((....))))))))...-........((..((((((((.......))))))))-))))).. ( -39.80) >consensus AGGGGAUGUUUUGGGCCAAAAAAGCUC_AACCCUCCCCAUCGACCUCAACAGGCCAACAGUUGAUGCUGUGCCAAAAAAAUAGUGGGAAGCACAAGGAAUUGAACUUUGUGC_CCGUCUG .(((((.(.((.((((.......)))).)).).))))).............(((..(((((....)))))))).......((((((...((((((((.......)))))))).))).))) (-34.41 = -34.73 + 0.31)

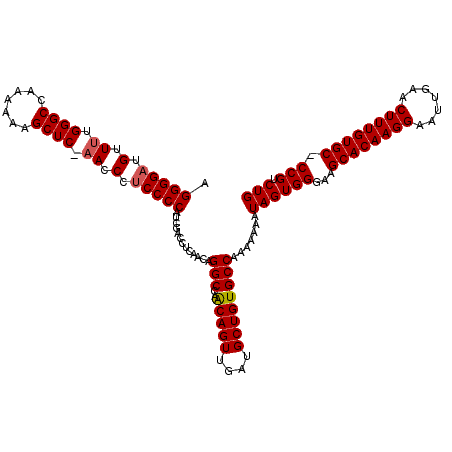
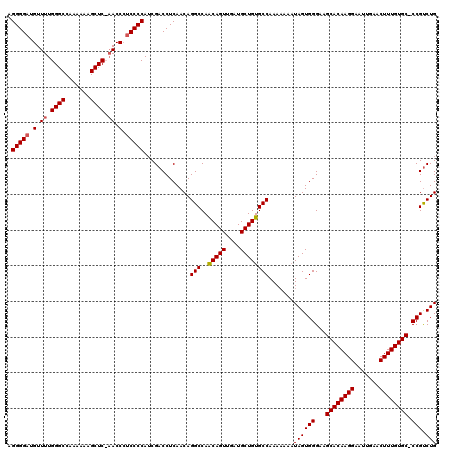
| Location | 14,030,502 – 14,030,622 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -34.88 |
| Energy contribution | -35.50 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14030502 120 - 20766785 CAGACGGGGCACAAAGUUCAAUUCCUUGUGCUUCCCACUAUUUUUUUGGCACAGCAUCAACUGUUGGCCUGUUGAGGUCGUUGGGGUGGGUUGGAGCUUUUUUGGCCCAAAACAUCCCCU ..((((((((((((.(.......).))))))..)))...........(((.(((((.....)))))))).......)))...((((((..((((.(((.....)))))))..)))))).. ( -44.00) >DroSim_CAF1 60584 119 - 1 CAGACGG-GCACAAAGUUCAAUUCCUUGUGCUUCCCACUAUUUUUUUGGCACAGCAUCAACUGUUGGCCUGUUGAGGUCGAUGGGGAGGGUUGGAGCUUUUUUGGCCCAAAACAUCCCCU ..(((((-((((((.(.......).))))))))..............(((.(((((.....)))))))).......)))...((((((..((((.(((.....)))))))..).))))). ( -40.10) >DroEre_CAF1 63816 117 - 1 CAGGCGG-GCACAAAGUUCAAUUCCUUGUGCUUCCCACUAUUUUUUUGGCACAGCAUCAACUGCUGGCCUGUUGAGGUCGAUGGGGAUGGUA-GAGCUUU-UUGGCCCAAAACAUCCCCU ..((..(-((((((.(.......).)))))))..)).(.(((((...(((.(((((.....))))))))....))))).)..(((((((...-(.(((..-..))).)....))))))). ( -40.80) >DroYak_CAF1 62456 117 - 1 CAGACGG-GCACAAAGUUCAAUUCCUUGUGCUACCCACUAAUU-UUUGGCACAGCAUCAACUGUUGGCCUGUUGAGGUCGAUGGGGAGGGUU-GAGCUUUUUUGGCCCAAAACAUCCCCU .....((-((..((((((((((.((((((((((..........-..))))))........((((((((((....)))))))))).)))))))-)))))))....))))............ ( -41.80) >consensus CAGACGG_GCACAAAGUUCAAUUCCUUGUGCUUCCCACUAUUUUUUUGGCACAGCAUCAACUGUUGGCCUGUUGAGGUCGAUGGGGAGGGUU_GAGCUUUUUUGGCCCAAAACAUCCCCU ..(((((.((((((.(.......).))))))...))...........(((.(((((.....)))))))).......)))...(((((...((((.(((.....)))))))....))))). (-34.88 = -35.50 + 0.62)

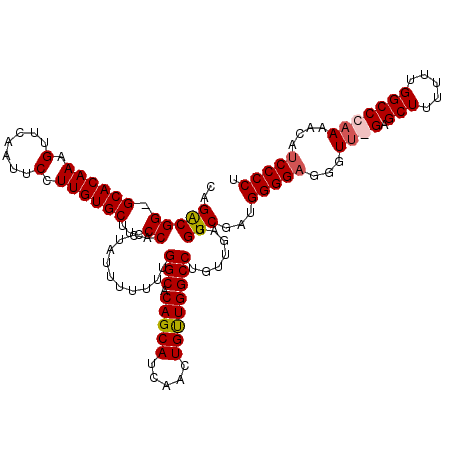
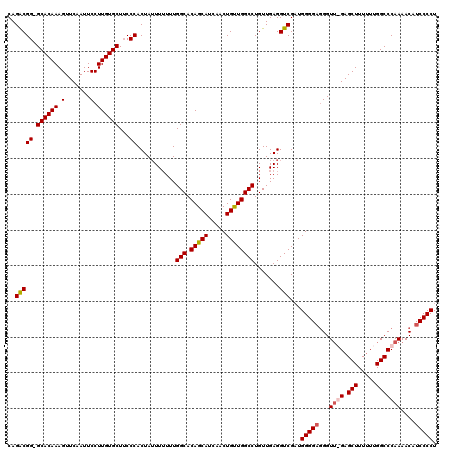
| Location | 14,030,582 – 14,030,696 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.42 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14030582 114 + 20766785 UAGUGGGAAGCACAAGGAAUUGAACUUUGUGCCCCGUCUGAUUGCACUUCGCUUCUUCCCUUCUCUA------UGUGUCUCUCUCGCACACUCUGUAGCCAUCUCUCUCGCUCUAGGGUG (((.(((((((((((((.......)))))))).......((..((.....)).)))))))....)))------((((.......))))(((((((.(((..........))).))))))) ( -29.40) >DroSim_CAF1 60664 113 + 1 UAGUGGGAAGCACAAGGAAUUGAACUUUGUGC-CCGUCUGAUUGCACUUCGCUUCCUCCCUUCUCUC------UCCGUCUCUCUCACACCCUAUGUAGCCAUCUCACUCGCUCUAGGGUG .((.((((((....(((((.((((....((((-..........)))))))).)))))..)))))).)------)............(((((((.(.(((..........))))))))))) ( -28.70) >DroEre_CAF1 63894 119 + 1 UAGUGGGAAGCACAAGGAAUUGAACUUUGUGC-CCGCCUGAUUGCACUUCUCUUCCCCCUCUCACCCCCUCCCGCCCUCUCUUUCCCACACUCUAUAGCCGUCUCUCUCGCUCUAGGGUG ..(((((((((((((((.......))))))))-..((......)).....................................)))))))((((((.(((..........))).)))))). ( -30.30) >consensus UAGUGGGAAGCACAAGGAAUUGAACUUUGUGC_CCGUCUGAUUGCACUUCGCUUCCUCCCUUCUCUC______UCCGUCUCUCUCACACACUCUGUAGCCAUCUCUCUCGCUCUAGGGUG ..((((((.((((((((.......))))))))...((......))....................................)))))).(((((((.(((..........))).))))))) (-21.20 = -21.43 + 0.23)

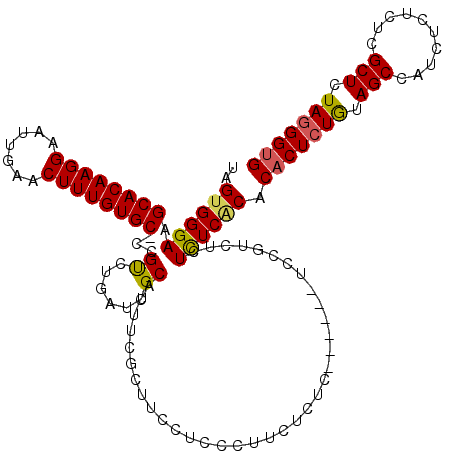
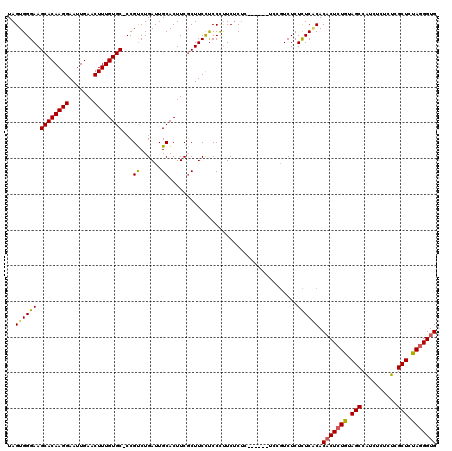
| Location | 14,030,622 – 14,030,736 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -20.44 |
| Energy contribution | -19.33 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14030622 114 - 20766785 UGUACAUAUAAGCCCAUUGUACAUAUCUAACUUUCGCUCUCACCCUAGAGCGAGAGAGAUGGCUACAGAGUGUGCGAGAGAGACACA------UAGAGAAGGGAAGAAGCGAAGUGCAAU (((((.......(((.(((((..(((((..((((((((((......)))))))))))))))..))))).((((.(....)..)))).------.......)))..........))))).. ( -35.23) >DroSim_CAF1 60703 110 - 1 UGU----AUAAGCCCAUUGUACAUAUCCAACUUUCGCUCUCACCCUAGAGCGAGUGAGAUGGCUACAUAGGGUGUGAGAGAGACGGA------GAGAGAAGGGAGGAAGCGAAGUGCAAU (((----((((.....)))))))..(((..(((((.(((((((((((.(((.(......).)))...))))))((.......)).))------))).)))))..))).((.....))... ( -35.80) >DroEre_CAF1 63933 116 - 1 UGU----AUAAGCCCAUUGUACAUAUUCAACUUUCGCUCUCACCCUAGAGCGAGAGAGACGGCUAUAGAGUGUGGGAAAGAGAGGGCGGGAGGGGGUGAGAGGGGGAAGAGAAGUGCAAU ...----........(((((((...(((...(..(.((((((((((....(....)...(.(((......(......)......))).)...)))))))))).)..)...)))))))))) ( -32.40) >consensus UGU____AUAAGCCCAUUGUACAUAUCCAACUUUCGCUCUCACCCUAGAGCGAGAGAGAUGGCUACAGAGUGUGCGAGAGAGACGGA______GAGAGAAGGGAGGAAGCGAAGUGCAAU ...............(((((((...(((..((((((((((......)))))..........((........))........................)))))..)))......))))))) (-20.44 = -19.33 + -1.10)

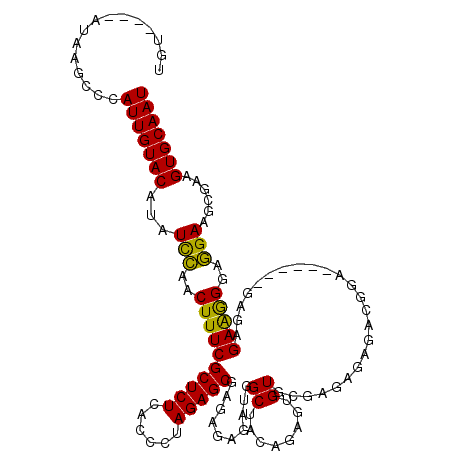
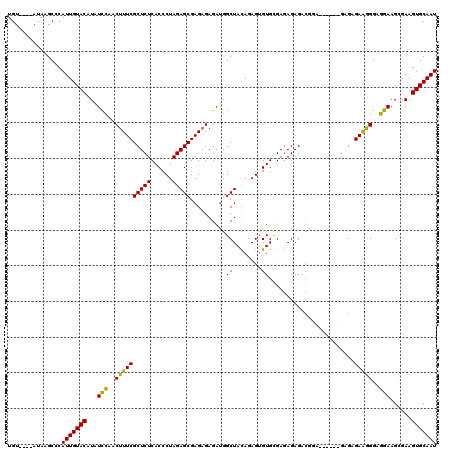
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:20 2006