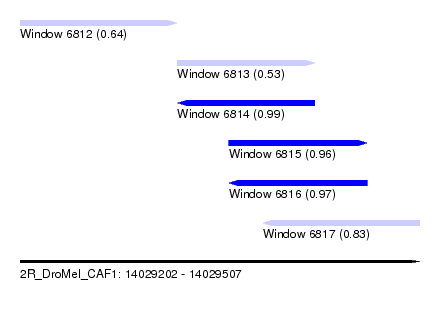
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,029,202 – 14,029,507 |
| Length | 305 |
| Max. P | 0.988282 |

| Location | 14,029,202 – 14,029,322 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -38.00 |
| Energy contribution | -37.84 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14029202 120 + 20766785 GCGGCGGCAUUUGAACUUUCAACCCCCGAAAGCCACGUGCUCCUGCAGCUUUUGGCCUCCUUUCCGCGUGCGGCAAUCAAUGAAAGUGAAGCUUUGCCCAAUUUAAGCCCGCUGCCAGCC ((((((((......(((((((....((((((((....(((....)))))))))))........(((....))).......)))))))...((((..........))))..)))))).)). ( -37.40) >DroSec_CAF1 59662 120 + 1 GCGGCGGCAUUUGAACUUUCAACCCCCGAAAGCCACGUGCUCCUGCAGCUUUUGGCCUCCUUUCCGCGUGCGGCAAUCAAUGAAAGUGAAGCUUUGCCCAAUUUAAGCCCGCUGCCAGCC ((((((((......(((((((....((((((((....(((....)))))))))))........(((....))).......)))))))...((((..........))))..)))))).)). ( -37.40) >DroSim_CAF1 59262 120 + 1 GCGGCGGCAUUUGAACUUUCAACCCCCGAAAGCCACGUGCUCCUGCAGCUUUUGGCCUCCUUUCCGCGUGCGGCAAUCAAUGAAAGUGAAGCUUUGCCCAAUUUAAGCCCGCUGCCAGCC ((((((((......(((((((....((((((((....(((....)))))))))))........(((....))).......)))))))...((((..........))))..)))))).)). ( -37.40) >DroEre_CAF1 62514 120 + 1 GCGGCGGCAUUUGAACUUUCAACCCCCGAAAGCCACGUGCUCCUGCAGCUUUUGGCCUCCUUUCCGCGUGCGGCAAUCAAUGAAAGUGAAGCUUUGCCCAAUUUAAGCCCGCCGCCAGCC ((((((((......(((((((....((((((((....(((....)))))))))))........(((....))).......)))))))...((((..........))))..)))))).)). ( -39.60) >DroYak_CAF1 61127 120 + 1 GCGGCGGCAUUUGAACUUUCAACCCCCGAAAGCCACGUGCUCCUGCAGCUUUUGGCCUCCUUUCCGCGUGCGGCAAUCAAUGAAAGUGAAGCUUUGCCCAAUUUAAGCCCGCUGCCAGCC ((((((((......(((((((....((((((((....(((....)))))))))))........(((....))).......)))))))...((((..........))))..)))))).)). ( -37.40) >consensus GCGGCGGCAUUUGAACUUUCAACCCCCGAAAGCCACGUGCUCCUGCAGCUUUUGGCCUCCUUUCCGCGUGCGGCAAUCAAUGAAAGUGAAGCUUUGCCCAAUUUAAGCCCGCUGCCAGCC ((((((((......(((((((....((((((((....(((....)))))))))))........(((....))).......)))))))...((((..........))))..)))))).)). (-38.00 = -37.84 + -0.16)


| Location | 14,029,322 – 14,029,427 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -28.39 |
| Energy contribution | -28.08 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14029322 105 + 20766785 CAAAAAUUUCCAUGCAGAGUCCUGCGUCAACGC-UUGACAUGUGUUCGU--------GUGUGUUUGUGUGUGUAGUGGAUGUUCCUGCGCUGGUGGUGGUUAGUUCACCACCGA ...........((((((....))))))..((((-..((((..(......--------)..)))).))))((((((.((....))))))))(((((((((.....))))))))). ( -34.20) >DroSec_CAF1 59782 97 + 1 CAAAAAUUUCCAUGCAGAGUCCUGCGUCAACGG-GUGACAUGUGUGCGU--------GUGU--------GUGUAGUGGAUGUUCCUGCGCUGGUGGUGGUUAGUUCACCACCGA ........(((((((((....))))))....))-)..(((((....)))--------))..--------((((((.((....))))))))(((((((((.....))))))))). ( -32.00) >DroSim_CAF1 59382 105 + 1 CAAAAAUUUCCAUGCAGAGUCCUGCGUCAACGG-UUGACAUGUGUGCGU--------GUGUGUUUGCGUGUGUAGUGGUUGUUCCUGCGUUGGUGGUGGUUAGUUCACCACCGA ((..(((..((((((((....))))((((....-.)))).....((((.--------.(((....)))..))))))))..)))..))..((((((((((.....)))))))))) ( -34.30) >DroEre_CAF1 62634 114 + 1 GAAAAAUUUCCAUGCAGAGUCCUGCGUCAACGCGUUGACGUGUGUAUGCAUGGGUGUGUGUGUGUGCGAGUGUAGUGGCUGUUUCUGCGCUGGUGGUGGUUAGUUCACCACCGA .............(((((((((((((((((....)))))(((..(((((((....)))))))..)))....)))).)))....)))))..(((((((((.....))))))))). ( -42.80) >consensus CAAAAAUUUCCAUGCAGAGUCCUGCGUCAACGC_UUGACAUGUGUGCGU________GUGUGUUUGCGUGUGUAGUGGAUGUUCCUGCGCUGGUGGUGGUUAGUUCACCACCGA ..........(((((((....))))(((((....))))))))...........................((((((.((....))))))))(((((((((.....))))))))). (-28.39 = -28.08 + -0.31)

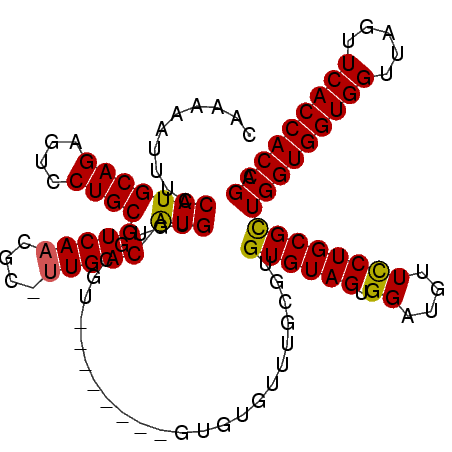
| Location | 14,029,322 – 14,029,427 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -25.46 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14029322 105 - 20766785 UCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCACUACACACACAAACACAC--------ACGAACACAUGUCAA-GCGUUGACGCAGGACUCUGCAUGGAAAUUUUUG ..(((((((.......)))))))....((((((..((((..................--------..........((((.-....))))((((....)))).))))..)))))) ( -27.70) >DroSec_CAF1 59782 97 - 1 UCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCACUACAC--------ACAC--------ACGCACACAUGUCAC-CCGUUGACGCAGGACUCUGCAUGGAAAUUUUUG ..(((((((.......)))))))....((((((..((((......--------....--------..........((((.-....))))((((....)))).))))..)))))) ( -27.60) >DroSim_CAF1 59382 105 - 1 UCGGUGGUGAACUAACCACCACCAACGCAGGAACAACCACUACACACGCAAACACAC--------ACGCACACAUGUCAA-CCGUUGACGCAGGACUCUGCAUGGAAAUUUUUG ..(((((((.......)))))))....((((((...(((........((........--------..))......((((.-....))))((((....)))).)))...)))))) ( -25.10) >DroEre_CAF1 62634 114 - 1 UCGGUGGUGAACUAACCACCACCAGCGCAGAAACAGCCACUACACUCGCACACACACACACCCAUGCAUACACACGUCAACGCGUUGACGCAGGACUCUGCAUGGAAAUUUUUC ..(((((((.......))))))).(((.((..............)))))............(((((((......((((((....))))))........)))))))......... ( -32.28) >consensus UCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCACUACACACGCAAACACAC________ACGCACACAUGUCAA_CCGUUGACGCAGGACUCUGCAUGGAAAUUUUUG ..(((((((.......)))))))....((((((..((((....................................(((((....)))))((((....)))).))))..)))))) (-25.46 = -26.28 + 0.81)

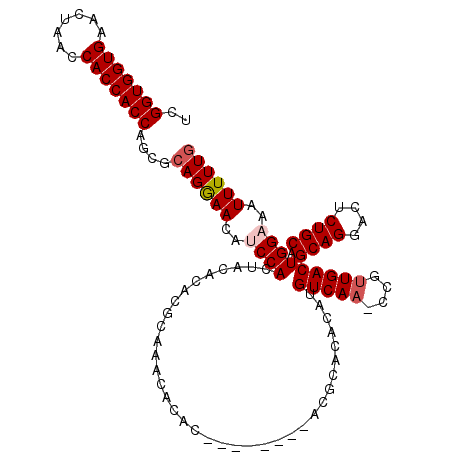

| Location | 14,029,361 – 14,029,467 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.03 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -33.69 |
| Energy contribution | -34.75 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14029361 106 + 20766785 UGUGUUCGU--------GUGUGUUUGUGUGUGUAGUGGAUGUUCCUGCGCUGGUGGUGGUUAGUUCACCACCGAUUCAAACACACCAUGCCAAUUCGCGAGCUUCGCAGUGCGG ...(((((.--------(((((((((.((((((((.((....))))))))(((((((((.....))))))))))).))))))))))..((......))))))..(((...))). ( -39.10) >DroSec_CAF1 59821 98 + 1 UGUGUGCGU--------GUGU--------GUGUAGUGGAUGUUCCUGCGCUGGUGGUGGUUAGUUCACCACCGAUUCAAACACACCAUGCCAAUUCGCGAGCUUCGCAGUGCGG .(((((.((--------((((--------((((((.((....))))))))(((((((((.....)))))))))......)))))))))))......(((.....)))....... ( -37.40) >DroSim_CAF1 59421 106 + 1 UGUGUGCGU--------GUGUGUUUGCGUGUGUAGUGGUUGUUCCUGCGUUGGUGGUGGUUAGUUCACCACCGAUUCAAACACACCAUGCCAAUUCGCGAGCUUCGCAGUGCGG ..((..(.(--------(((.(((((((..((..(((((((((..((.(((((((((((.....))))))))))).)))))).)))))..))...)))))))..)))))..)). ( -46.30) >DroEre_CAF1 62674 114 + 1 UGUGUAUGCAUGGGUGUGUGUGUGUGCGAGUGUAGUGGCUGUUUCUGCGCUGGUGGUGGUUAGUUCACCACCGAUCCAAACACACCAUGCCAAUUCGCGAGCUUCGCAGUGCGG ......(((((.((((((.((((((...(((((((.........)))))))((((((((.....)))))))).......)))))))))))).....(((.....))).))))). ( -47.20) >consensus UGUGUGCGU________GUGUGUUUGCGUGUGUAGUGGAUGUUCCUGCGCUGGUGGUGGUUAGUUCACCACCGAUUCAAACACACCAUGCCAAUUCGCGAGCUUCGCAGUGCGG ..(((((..........(((((((((...((((((.((....))))))))(((((((((.....)))))))))...)))))))))...........(((.....))).))))). (-33.69 = -34.75 + 1.06)

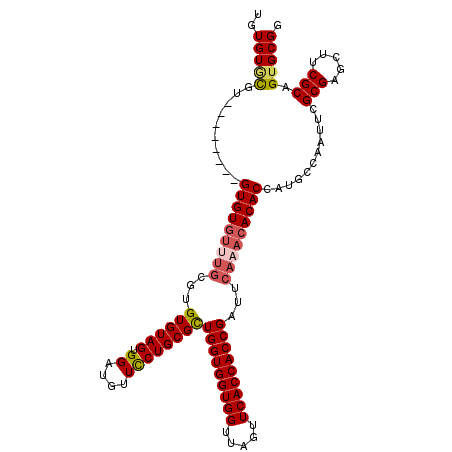
| Location | 14,029,361 – 14,029,467 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.03 |
| Mean single sequence MFE | -34.09 |
| Consensus MFE | -25.95 |
| Energy contribution | -27.70 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14029361 106 - 20766785 CCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGAAUCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCACUACACACACAAACACAC--------ACGAACACA ..((..((((.....)))).....))...(((((((((....(((((((.......)))))))......((.....))..........)))))))))--------......... ( -33.70) >DroSec_CAF1 59821 98 - 1 CCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGAAUCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCACUACAC--------ACAC--------ACGCACACA ..((..((((.....))))..........((((((.((....(((((((.......)))))))......((.....))....)).--------))))--------))))..... ( -31.30) >DroSim_CAF1 59421 106 - 1 CCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGAAUCGGUGGUGAACUAACCACCACCAACGCAGGAACAACCACUACACACGCAAACACAC--------ACGCACACA ..((..((((.....))))..........(((((((((....(((((((.......)))))))......((.....))..........)))))))))--------..))..... ( -33.50) >DroEre_CAF1 62674 114 - 1 CCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGGAUCGGUGGUGAACUAACCACCACCAGCGCAGAAACAGCCACUACACUCGCACACACACACACCCAUGCAUACACA ......((((.....)))).....(((((((((((.((....(((((((.......))))))).(((.((..............)))))...)).))))).))))))....... ( -37.84) >consensus CCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGAAUCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCACUACACACGCAAACACAC________ACGCACACA ..((..((((.....)))).....))...(((((((((....(((((((.......)))))))......((.....))..........)))))))))................. (-25.95 = -27.70 + 1.75)

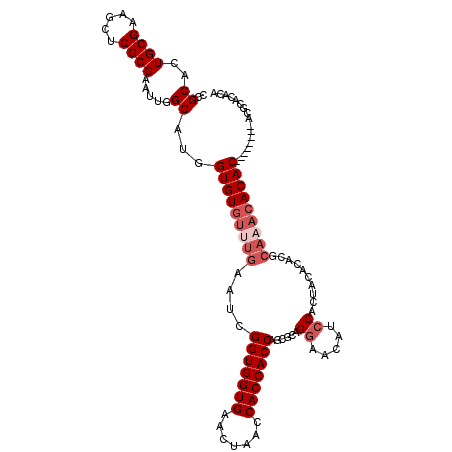
| Location | 14,029,387 – 14,029,507 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.58 |
| Mean single sequence MFE | -39.84 |
| Consensus MFE | -36.38 |
| Energy contribution | -36.78 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14029387 120 - 20766785 UUGUGCUUUCCGUUGUAAACUGAAUCCAAAUCGCUUGGCGCCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGAAUCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCAC .((((((....(...(((((...((((((.((((..(((..(((...)))..))).)))).)))).)).....)))))...)(((((((.......)))))))))))))((.....)).. ( -40.10) >DroSec_CAF1 59839 120 - 1 UUGUGCUUUCCGUUGUAAACUGAAUCCAAAUCGCUUGGCGCCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGAAUCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCAC .((((((....(...(((((...((((((.((((..(((..(((...)))..))).)))).)))).)).....)))))...)(((((((.......)))))))))))))((.....)).. ( -40.10) >DroSim_CAF1 59447 120 - 1 UUGUGCUUUCCGUUGUAAACUGAAUCCAAAUCGCUUGGCGCCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGAAUCGGUGGUGAACUAACCACCACCAACGCAGGAACAACCAC ((((.(((..((((((((((...((((((.((((..(((..(((...)))..))).)))).)))).)).....)))))....((((((......))))))..))))).))).)))).... ( -36.80) >DroEre_CAF1 62708 120 - 1 UUGUGCUUUCCGUUGUAAACUAAAUCCAAAUCGCUUGGCGCCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGGAUCGGUGGUGAACUAACCACCACCAGCGCAGAAACAGCCAC (((((((.((((......((((...((((.((((..(((..(((...)))..))).)))).))))..)))).....))))..(((((((.......)))))))))))))).......... ( -42.20) >DroYak_CAF1 61327 120 - 1 UUGUGCUUUCCGUUGUAAACUAAACCCAAAUCGCUUGGCGCCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGAAUCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCAC .((((((....(...(((((...((((((.((((..(((..(((...)))..))).)))).)))....)))..)))))...)(((((((.......)))))))))))))((.....)).. ( -40.00) >consensus UUGUGCUUUCCGUUGUAAACUGAAUCCAAAUCGCUUGGCGCCGCACUGCGAAGCUCGCGAAUUGGCAUGGUGUGUUUGAAUCGGUGGUGAACUAACCACCACCAGCGCAGGAACAUCCAC (((((((....(...(((((...((((((.((((..(((..(((...)))..))).)))).)))).)).....)))))...)(((((((.......)))))))))))))).......... (-36.38 = -36.78 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:12 2006