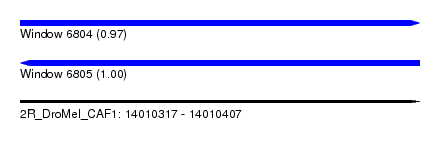
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,010,317 – 14,010,407 |
| Length | 90 |
| Max. P | 0.995535 |

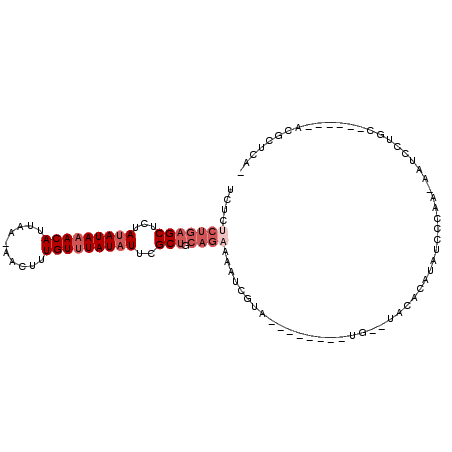
| Location | 14,010,317 – 14,010,407 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.66 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -11.98 |
| Energy contribution | -12.82 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14010317 90 + 20766785 -UGAGCGU------GCAGGAUU-UUGGGAUAUGUGUA--CA--------UACGAUUUUCUGCAGCGAAUAUAAACAAAGUU-UUAAUGUUUAUAUAGAGCUCAGAGAGA -(((((.(------((((((..-(((...((((....--))--------)))))..)))))))....(((((((((.....-....)))))))))...)))))...... ( -25.10) >DroSec_CAF1 40997 90 + 1 -UGAGCGU------GCUGUAUU-UUGGGAUAUACGUA--CA--------UACGAUUUUCUGCAGCGAAUAUAAACAAAGUU-UUAAUGUUUAUAUAGAGCUCAGAGAGA -.....((------((((((((-....)))))).)))--).--------.....(((((((.(((..(((((((((.....-....)))))))))...)))))))))). ( -23.80) >DroSim_CAF1 40385 90 + 1 -UGAGCGU------GCAGGAUU-UUGGGAUAUGCGUA--CA--------UACGAUAUUCUGCAGCGAAUAUAAACAAAGUU-UUAAUGUUUAUAUAGAGCUCAGAGAGA -(((((.(------(((((((.-(((...((((....--))--------))))).))))))))....(((((((((.....-....)))))))))...)))))...... ( -24.50) >DroEre_CAF1 41517 97 + 1 -UGAGCGU------GCAGGAUU-UUGGGAUAUGUGUA--CAUA--CUCGUACGAUUUUCUGCAGCGAAUAUAAACAAAGUUUUUAAUGUUUAUAUAGAGCUCAGAGAGA -(((((.(------((((((..-(((.((((((....--))))--.))...)))..)))))))....(((((((((..........)))))))))...)))))...... ( -26.80) >DroYak_CAF1 41108 103 + 1 GUGAGCGU------GCAGGAUUUUUGGGAUGUGUGUGUGUGUACACAUACACAAUUUGCUGCAGCGAAUAUAAACAAAGUUUUUAAUGUUUAUAUAGAGCUCAGAGAGA .(((((.(------((((.(..(......((((((((((....)))))))))))..).)))))....(((((((((..........)))))))))...)))))...... ( -31.90) >DroAna_CAF1 39970 87 + 1 -UGAGUGUUGAUAUAAAAGAUU-UUG-GAUUUCUGGU--CG--------AA-GAAUCGCGCGUGCAAAUAUAAACAAUGUU-UU-AUGUGUAUUUACAGCUCA------ -.((((((.(((......((((-((.-(((.....))--).--------.)-)))))(((((((.((((((.....)))))-))-))))))))).))).))).------ ( -19.20) >consensus _UGAGCGU______GCAGGAUU_UUGGGAUAUGUGUA__CA________UACGAUUUUCUGCAGCGAAUAUAAACAAAGUU_UUAAUGUUUAUAUAGAGCUCAGAGAGA .(((((....................(((....((((((........))))))....))).......(((((((((..........)))))))))...)))))...... (-11.98 = -12.82 + 0.84)

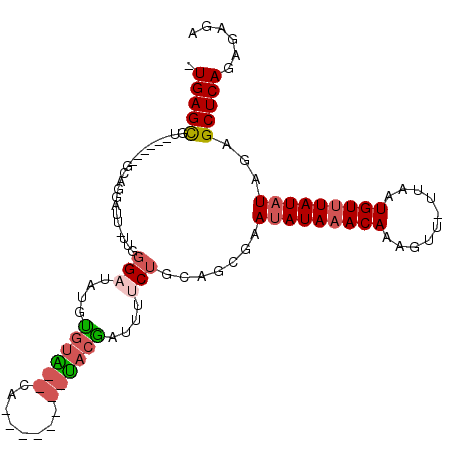
| Location | 14,010,317 – 14,010,407 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.66 |
| Mean single sequence MFE | -19.17 |
| Consensus MFE | -9.83 |
| Energy contribution | -11.17 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14010317 90 - 20766785 UCUCUCUGAGCUCUAUAUAAACAUUAA-AACUUUGUUUAUAUUCGCUGCAGAAAAUCGUA--------UG--UACACAUAUCCCAA-AAUCCUGC------ACGCUCA- .......((((...(((((((((....-.....)))))))))..(.(((((......(((--------((--....))))).....-....))))------)))))).- ( -19.06) >DroSec_CAF1 40997 90 - 1 UCUCUCUGAGCUCUAUAUAAACAUUAA-AACUUUGUUUAUAUUCGCUGCAGAAAAUCGUA--------UG--UACGUAUAUCCCAA-AAUACAGC------ACGCUCA- ....(((((((...(((((((((....-.....)))))))))..))).)))).....(..--------((--(..((((.......-.)))).))------)..)...- ( -18.30) >DroSim_CAF1 40385 90 - 1 UCUCUCUGAGCUCUAUAUAAACAUUAA-AACUUUGUUUAUAUUCGCUGCAGAAUAUCGUA--------UG--UACGCAUAUCCCAA-AAUCCUGC------ACGCUCA- .......((((...(((((((((....-.....)))))))))..(.(((((......(((--------((--....))))).....-....))))------)))))).- ( -19.36) >DroEre_CAF1 41517 97 - 1 UCUCUCUGAGCUCUAUAUAAACAUUAAAAACUUUGUUUAUAUUCGCUGCAGAAAAUCGUACGAG--UAUG--UACACAUAUCCCAA-AAUCCUGC------ACGCUCA- ....(((((((...(((((((((..........)))))))))..))).)))).........(((--..((--((............-.....)))------)..))).- ( -20.83) >DroYak_CAF1 41108 103 - 1 UCUCUCUGAGCUCUAUAUAAACAUUAAAAACUUUGUUUAUAUUCGCUGCAGCAAAUUGUGUAUGUGUACACACACACACAUCCCAAAAAUCCUGC------ACGCUCAC ......(((((...(((((((((..........)))))))))..(.(((((.....(((((.((((....)))).)))))...........))))------))))))). ( -25.99) >DroAna_CAF1 39970 87 - 1 ------UGAGCUGUAAAUACACAU-AA-AACAUUGUUUAUAUUUGCACGCGCGAUUC-UU--------CG--ACCAGAAAUC-CAA-AAUCUUUUAUAUCAACACUCA- ------(((((((((((((.(((.-..-.....)))...)))))))).)).(((...-.)--------))--..........-...-...........))).......- ( -11.50) >consensus UCUCUCUGAGCUCUAUAUAAACAUUAA_AACUUUGUUUAUAUUCGCUGCAGAAAAUCGUA________UG__UACACAUAUCCCAA_AAUCCUGC______ACGCUCA_ ....(((((((...(((((((((..........)))))))))..))).))))......................................................... ( -9.83 = -11.17 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:00 2006