| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,984,226 – 13,984,327 |
| Length | 101 |
| Max. P | 0.681175 |

| Location | 13,984,226 – 13,984,327 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -18.89 |
| Energy contribution | -18.87 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
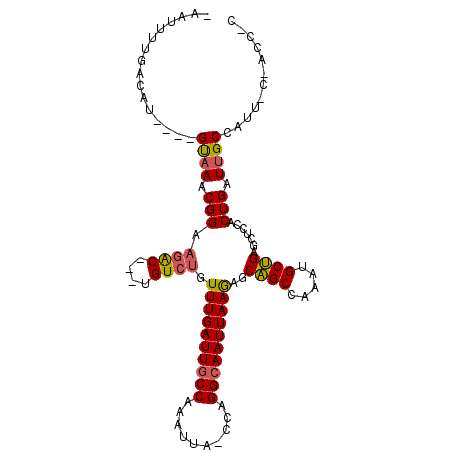
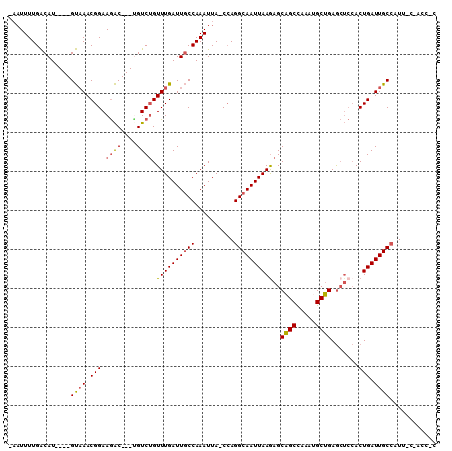
>2R_DroMel_CAF1 13984226 101 + 20766785 -AAUUUUGACAU----GUAAACGGAAGAC---UGUCUGUUUGAUUGCCAAAUUA-CCAGGCAAUUAAGAGCAGCCAAACGCUGAGCUCCACUGAUUGCCUUCCCCACGCC -...((((.((.----.((((((((....---..))))))))..)).))))...-..(((((((((.(((((((.....)))..))))...))))))))).......... ( -26.60) >DroSec_CAF1 22540 101 + 1 -UAUUUUGACAU----GUAAACGGAAGAC---UGUCUGUUUGAUUGCCAAAUUA-CCAGGCAAUUAAGAGCAGCCAAAUGCUGAGCUCCACUGAUUGCCAUUUCGACCUC -...((((.((.----.((((((((....---..))))))))..)).))))...-...((((((((.(((((((.....)))..))))...))))))))........... ( -26.40) >DroSim_CAF1 22199 101 + 1 -AAUUUUGACAU----GUAAACGGAAGAC---UGUCUGUUUGAUUGCCAAAUUA-CCAGGCAAUUAAGAGCAGCCAAAUGCUGAGCUCCACUGAUUGCCAUUUCGACCCC -...((((.((.----.((((((((....---..))))))))..)).))))...-...((((((((.(((((((.....)))..))))...))))))))........... ( -26.40) >DroEre_CAF1 22981 94 + 1 -AAUUUUGACAU----GUAAACGGAAGAC---CGUCUGUUUGAUUGCCAAAUUA-CCAGGCAAUUAAGAGCAGCCAAAUGCUGAGCUCCACUGAUUGCCUUU-------C -...((((.((.----.((((((((....---..))))))))..)).))))...-..(((((((((.(((((((.....)))..))))...)))))))))..-------. ( -26.50) >DroYak_CAF1 22731 94 + 1 -AAUUUUGACAU----GUAAACGGAAGAC---GGUCUGUUUGAUUGCCAAAUUA-CGAGGCAAUUAAAAGCAGCCCAAUGCUGAGCUCCACUGAUUGCCAUU-------C -...((((.((.----.((((((((....---..))))))))..)).))))...-...((((((((..((((((.....)))..)))....))))))))...-------. ( -23.00) >DroAna_CAF1 22642 104 + 1 CAUUUUUGGCAUGACUGCCAACGGAAAGCGGUUGUCAGUUUGAUUUCCAAAUUAGCCAGGCAAUUAAGAGCGGCUCAUGGCCGAA----ACUGAUUGCAUUU-CCAGCU- .....((((((....)))))).(((((..(((((...(((((.....))))))))))..(((((((....((((.....))))..----..))))))).)))-))....- ( -33.50) >consensus _AAUUUUGACAU____GUAAACGGAAGAC___UGUCUGUUUGAUUGCCAAAUUA_CCAGGCAAUUAAGAGCAGCCAAAUGCUGAGCUCCACUGAUUGCCAUU_C_ACC_C ................((((.(((.((((....)))).((((((((((..........))))))))))..((((.....)))).......))).))))............ (-18.89 = -18.87 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:58 2006