
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,981,591 – 13,981,710 |
| Length | 119 |
| Max. P | 0.773396 |

| Location | 13,981,591 – 13,981,710 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -21.01 |
| Energy contribution | -22.05 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
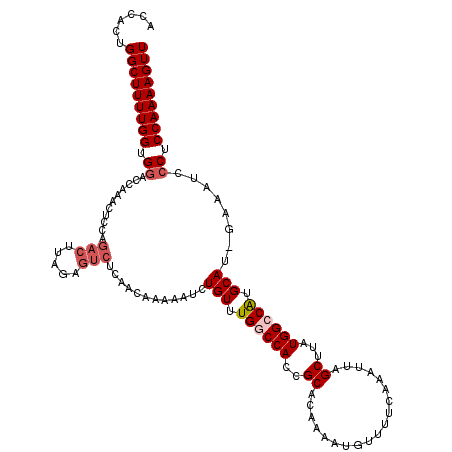
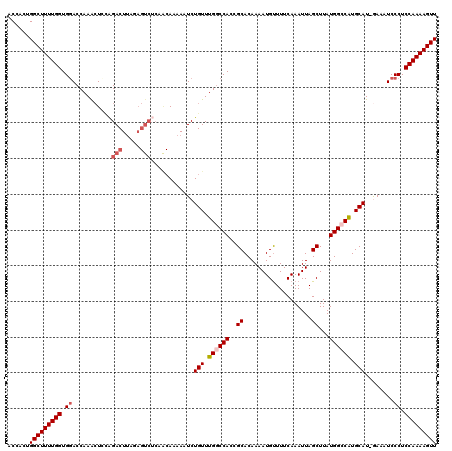
>2R_DroMel_CAF1 13981591 119 + 20766785 ACCACUGGCUUUUGGUGGACCAAACUUCAGACUUAUAGUCUCAACAAAAAUCUGUUUGGCCACCGCACAAAAUGUUUUCAAAUUAGCUUAUGGACAUGCAU-GAAAUCCCUCCAAAAGUU .(((..((((...(((((.((((((...((((.....))))............)))))))))))(((.....))).........))))..)))........-.................. ( -25.56) >DroSec_CAF1 19896 119 + 1 ACCAUGGGCUUUUGGUGGACCAAGCUCCAGACUUACAGUCUCAGCAAAAAUCUGUUUGGCCACCGCACAAAAUGUUUUCAAAUUAGCUUAUGGCCAUGCAU-GAAAUCCCUCCAAAAGUU ......(((((((((.((.....(((..((((.....)))).))).....(((((.((((((..((...................))...)))))).))).-))....)).))))))))) ( -32.81) >DroSim_CAF1 18995 119 + 1 ACCACGGGCUUUUGGUGGACCAAACUCCAGACUUAGAGUCUCAACAAAAAUCUGUUUGGCCACCGCACAAAAUGUUUUCAAAUUAGCUUAUGGCCGUGCAU-GAAAUCCCUCCAAAAGUU .....(((.(((((((((.((((((...((((.....))))............)))))))))))((((....((....)).....((.....)).))))..-)))).))).......... ( -30.66) >DroEre_CAF1 20297 108 + 1 ACACAUGGCUUUUGGUGGACCAAAAUGUGGACU------------AAAAAUCUGUUUGACCAACGCACAAAAUGCUUUCAAAUUAGCUUAUGGCCAUGCAUGAAAAUCCCUCCAAAAGUU ......(((((((((.(((.....((((((.((------------(.......((((((.....(((.....)))..)))))).......)))))..)))).....)))..))))))))) ( -22.74) >DroYak_CAF1 20319 119 + 1 ACCACUGGCUUUUGGUGUACCAAACUCGGGACUUAGAGUCGCAACAAAAAUCUGUUUGGCCAACGCACAAAAUGUUUUCAAAUUAGCUUAUGGCCAUGCAU-AAAAUCCCUCCAAAAGUU ......(((((((((.(..........(.(((.....))).)..........(((.((((((..((...................))...)))))).))).-.......).))))))))) ( -25.91) >consensus ACCACUGGCUUUUGGUGGACCAAACUCCAGACUUAGAGUCUCAACAAAAAUCUGUUUGGCCACCGCACAAAAUGUUUUCAAAUUAGCUUAUGGCCAUGCAU_GAAAUCCCUCCAAAAGUU ......(((((((((.((...........(((.....)))............(((.((((((..((...................))...)))))).)))........)).))))))))) (-21.01 = -22.05 + 1.04)

| Location | 13,981,591 – 13,981,710 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -24.60 |
| Energy contribution | -26.64 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
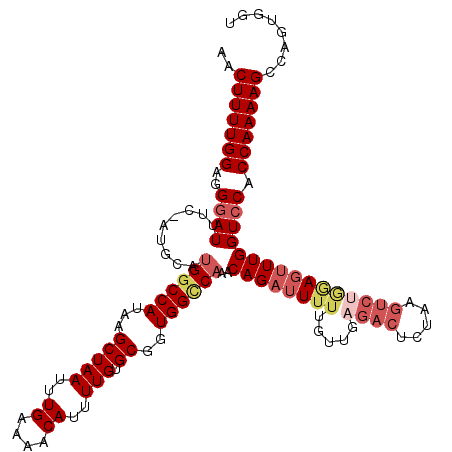
>2R_DroMel_CAF1 13981591 119 - 20766785 AACUUUUGGAGGGAUUUC-AUGCAUGUCCAUAAGCUAAUUUGAAAACAUUUUGUGCGGUGGCCAAACAGAUUUUUGUUGAGACUAUAAGUCUGAAGUUUGGUCCACCAAAAGCCAGUGGU .(((.((((..((((...-......))))....(((((..((....))..))).))(((((((((((((((((..((....))...))))))...)))))).))))).....)))).))) ( -31.50) >DroSec_CAF1 19896 119 - 1 AACUUUUGGAGGGAUUUC-AUGCAUGGCCAUAAGCUAAUUUGAAAACAUUUUGUGCGGUGGCCAAACAGAUUUUUGCUGAGACUGUAAGUCUGGAGCUUGGUCCACCAAAAGCCCAUGGU ..(((((((..(((((..-..((.(((((((..(((((..((....))..))).)).)))))))..(((((..((((.......)))))))))..))..))))).)))))))........ ( -36.80) >DroSim_CAF1 18995 119 - 1 AACUUUUGGAGGGAUUUC-AUGCACGGCCAUAAGCUAAUUUGAAAACAUUUUGUGCGGUGGCCAAACAGAUUUUUGUUGAGACUCUAAGUCUGGAGUUUGGUCCACCAAAAGCCCGUGGU ..(((((((..(((((..-..((..((((((..(((((..((....))..))).)).))))))...(((((((..((....))...)))))))..))..))))).)))))))........ ( -34.80) >DroEre_CAF1 20297 108 - 1 AACUUUUGGAGGGAUUUUCAUGCAUGGCCAUAAGCUAAUUUGAAAGCAUUUUGUGCGUUGGUCAAACAGAUUUUU------------AGUCCACAUUUUGGUCCACCAAAAGCCAUGUGU ..(((((((..(((((...(((..((((((...(((((..((....))..))).))..))))))....(((....------------.)))..)))...))))).)))))))........ ( -25.90) >DroYak_CAF1 20319 119 - 1 AACUUUUGGAGGGAUUUU-AUGCAUGGCCAUAAGCUAAUUUGAAAACAUUUUGUGCGUUGGCCAAACAGAUUUUUGUUGCGACUCUAAGUCCCGAGUUUGGUACACCAAAAGCCAGUGGU .(((.((((.(((((((.-.(((.((((((...(((((..((....))..))).))..))))))(((((....)))))))).....)))))))...(((((....)))))..)))).))) ( -35.60) >consensus AACUUUUGGAGGGAUUUC_AUGCAUGGCCAUAAGCUAAUUUGAAAACAUUUUGUGCGGUGGCCAAACAGAUUUUUGUUGAGACUCUAAGUCUGGAGUUUGGUCCACCAAAAGCCAGUGGU ..(((((((..((((.........((((((...(((((..((....))..))).))..))))))..((((((((.....((((.....)))))))))))))))).)))))))........ (-24.60 = -26.64 + 2.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:57 2006