
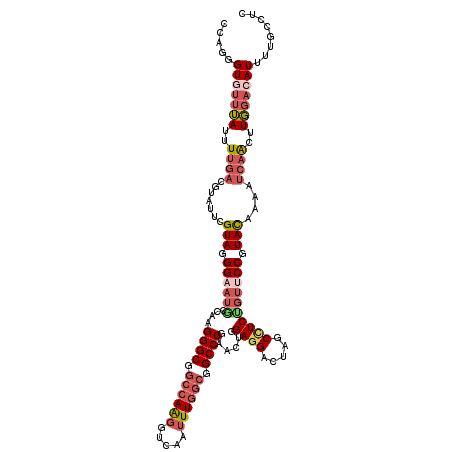
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,922,717 – 13,922,872 |
| Length | 155 |
| Max. P | 0.768694 |

| Location | 13,922,717 – 13,922,834 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -42.46 |
| Consensus MFE | -28.75 |
| Energy contribution | -30.37 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13922717 117 - 20766785 CCAGGGUGUUUAUUUUGACGUAUUCGUAGGGAAUGCCAAGGGCGGCCAAGGUCAAUUUGGCGGCCCGAACUGGAGGACUAGCCUCUGUCCCGUACAAAAUCAGCUUGGACAU---GCCGU (((((.((...(((((((((....))).((((.......((((.((((((.....)))))).)))).....(((((.....))))).))))...)))))))).)))))....---..... ( -44.30) >DroSec_CAF1 210379 120 - 1 CCAGGGUGUUUAUUUUGACGUAUUCGUAGGGAAUGCCAAGGGCGGCCAAGGUCAGUUUGGCGGCCCGAACUGGAGGACUAGCCUCAGUUCCGUACAAAAUCAAUUUGGACAUUUUGCCCU .(((((((((((..((((.((((((.....))))))...((((.((((((.....)))))).))))((((((.(((.....))))))))).........))))..))))))))))).... ( -46.80) >DroSim_CAF1 212838 120 - 1 CCAGGGUGUUUAUUUUGACGUAUUCGUAGGGAAUGCCAAGGGCGGCCAAGGUCAGUUUGGCGGCCCGAACUGGAGGACUAGCCUCUGUUCCGUACAAAAUCAACUUGGACAUUUUGCCUU .(((((((((((..((((.((((((.....))))))...((((.((((((.....)))))).))))((((.(((((.....))))))))).........))))..))))))))))).... ( -45.30) >DroEre_CAF1 210712 120 - 1 CCCGGGUAUUUAUUUUGACGUAUUCGUAGGGAAUACCGAGGGCGCCCAAGGUCAAUUUGGCGGCCCGAACUGGAGGACUAGCCUCUGUUCCGUACAAAAUCAACUUGGACAUAUUGCCUC .((((((....((((((((((((((.....))))))...((((.((((((.....))))).)))))((((.(((((.....))))))))).)).))))))..))))))............ ( -38.10) >DroYak_CAF1 222654 120 - 1 CCAGGGUAUUUAUUUUGACAUAUUCGUAGGGAAUACCAAGGGCGGCCAAGGUCAAUUUGGCGGCCCGCACCGGAGGACUAGCUUCUGUUCCGUACAAAAUCAACUUGGACAUUAUGCCCC ...((((((....((..(.......(((.((((......((((.((((((.....)))))).))))....((((((.....)))))))))).))).........)..))....)))))). ( -39.59) >DroAna_CAF1 222363 120 - 1 UCCUGGUGUUGACUUCUACGAUUUCGUAGGGAACUCCGAGGGCUGCCAGCGUAAGCUUUACGGCCCUAAAAGGAGGACUGUCAUCAAAACCAUAUAGUACUAGUUUUCCCAUGUUAGCUC .......((((((...((((....))))(((((((((.(((((((..(((....)))...)))))))....)))).(((((............))))).......)))))..)))))).. ( -40.70) >consensus CCAGGGUGUUUAUUUUGACGUAUUCGUAGGGAAUGCCAAGGGCGGCCAAGGUCAAUUUGGCGGCCCGAACUGGAGGACUAGCCUCUGUUCCGUACAAAAUCAACUUGGACAUUUUGCCUC .....(((((((..((((.......(((.((((((....((((.((((((.....)))))).))))......((((.....)))))))))).)))....))))..)))))))........ (-28.75 = -30.37 + 1.61)

| Location | 13,922,754 – 13,922,872 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.66 |
| Mean single sequence MFE | -44.68 |
| Consensus MFE | -33.44 |
| Energy contribution | -33.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13922754 118 - 20766785 AUUCUUCCGCAAGAACUCGGGCGA--AAGGGUCUCCUUGGCCAGGGUGUUUAUUUUGACGUAUUCGUAGGGAAUGCCAAGGGCGGCCAAGGUCAAUUUGGCGGCCCGAACUGGAGGACUA ..((((((......((((.(((..--((((....)))).))).))))((((........((((((.....))))))...((((.((((((.....)))))).)))))))).))))))... ( -46.40) >DroSec_CAF1 210419 118 - 1 AUUCUUUCGCAAGAACUCGGGCGA--AAGGGUCUCCUUGGCCAGGGUGUUUAUUUUGACGUAUUCGUAGGGAAUGCCAAGGGCGGCCAAGGUCAGUUUGGCGGCCCGAACUGGAGGACUA ...(((((((..(....)..))))--)))(((((((((((((..(((((((......(((....)))...)))))))......))))))))(((((((((....))))))))).))))). ( -48.20) >DroSim_CAF1 212878 118 - 1 AUUCUUCCGCACGAACUCGGGCGA--AAGGGUCUCCUUGGCCAGGGUGUUUAUUUUGACGUAUUCGUAGGGAAUGCCAAGGGCGGCCAAGGUCAGUUUGGCGGCCCGAACUGGAGGACUA ..((((((......((((.(((..--((((....)))).))).))))((((........((((((.....))))))...((((.((((((.....)))))).)))))))).))))))... ( -45.80) >DroEre_CAF1 210752 118 - 1 AUUCUUUCGCAAGAACUCAGGCGA--AAGGGUCUCCUUGGCCCGGGUAUUUAUUUUGACGUAUUCGUAGGGAAUACCGAGGGCGCCCAAGGUCAAUUUGGCGGCCCGAACUGGAGGACUA ...(((((((..(....)..))))--)))((((((((((((((.(((((((......(((....)))...)))))))..))))...)))))(((.(((((....))))).))).))))). ( -41.40) >DroYak_CAF1 222694 118 - 1 AUUCUUGCGCAGGAACUCAGGCGA--AAGGGUCUCCUUUGCCAGGGUAUUUAUUUUGACAUAUUCGUAGGGAAUACCAAGGGCGGCCAAGGUCAAUUUGGCGGCCCGCACCGGAGGACUA ...(((.(((..(....)..))).--)))(((((((..(((...(((((((....(((.....)))....)))))))..((((.((((((.....)))))).)))))))..))).)))). ( -45.00) >DroAna_CAF1 222403 118 - 1 AUUUCUCUUCAGGUAUUCGGGUUUAUAAUGG--UCCUUGUUCCUGGUGUUGACUUCUACGAUUUCGUAGGGAACUCCGAGGGCUGCCAGCGUAAGCUUUACGGCCCUAAAAGGAGGACUG ........(((((.((..((..(......).--.))..)).))))).(((...(((((((....)))))))..((((.(((((((..(((....)))...)))))))....))))))).. ( -41.30) >consensus AUUCUUCCGCAAGAACUCGGGCGA__AAGGGUCUCCUUGGCCAGGGUGUUUAUUUUGACGUAUUCGUAGGGAAUGCCAAGGGCGGCCAAGGUCAAUUUGGCGGCCCGAACUGGAGGACUA ..(((((((.....((((.(((....((((....)))).))).))))............((((((.....))))))...((((.((((((.....)))))).))))....)))))))... (-33.44 = -33.28 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:50 2006