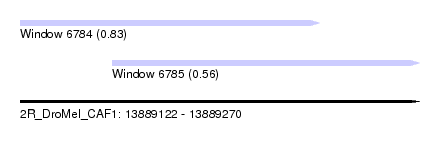
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,889,122 – 13,889,270 |
| Length | 148 |
| Max. P | 0.834410 |

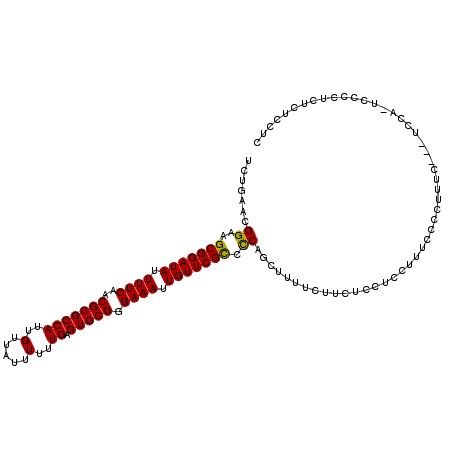
| Location | 13,889,122 – 13,889,233 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.62 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13889122 111 + 20766785 ---UCUUUCUGCUCGGG---UAUUUUCCUUUUGGUAGCUUUCUGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGCCCCAGCUUUUCUU ---.......(((.(((---....((((.((..(.......)..)).))))(((((((.((((..(((((((..(.....)..)).))))).)))).)))))))))))))....... ( -33.20) >DroPse_CAF1 171627 109 + 1 -------UCUGCUCUGGCGCUGUUUUCCUUUCGGUAGCUUUCGGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGUCUCAUCUCGCAU- -------...((..((((((.((((((.((((((......)))))).))))))(((....)))...))))))..))..........((((((((...((.......))))))))))- ( -32.50) >DroSim_CAF1 178017 97 + 1 --------------------UAUUUUCCUUUUGGUAGCUUUCUGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGCCCCAGCUUUUCUU --------------------....((((.((..(.......)..)).))))(((((((.((((..(((((((..(.....)..)).))))).)))).)))))))............. ( -25.30) >DroWil_CAF1 213130 105 + 1 CUUUUAUUUUUUUU--G---UGUU---UUUUUGGUAGCUUUCCAAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGUCGCAACGUU---- ............((--(---((((---..(((((.......)))))..)).(((((((.((((..(((((((..(.....)..)).))))).)))).))))))))))))....---- ( -24.40) >DroAna_CAF1 181191 107 + 1 ---UCUUUCGGCUUGGG---UAUUUUCCUUUUGGUAGCUUUCGGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCAUCCCACCUCU---- ---......((..((..---((.(((((........(((((((...)))))))............(((((((..(.....)..)).)))))))))).))..))..))......---- ( -29.20) >DroPer_CAF1 169050 109 + 1 -------UCUGCUCUGGCGCUGUUUUCCUUUCGGUAGCUUUCGGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGUCUCAUCUCGCAU- -------...((..((((((.((((((.((((((......)))))).))))))(((....)))...))))))..))..........((((((((...((.......))))))))))- ( -32.50) >consensus _______UCUGCUC_GG___UAUUUUCCUUUUGGUAGCUUUCGGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGUCCCAGCUCU__U_ ....................................((((((.....))))))(((((.((((..(((((((..(.....)..)).))))).)))).)))))............... (-21.62 = -21.62 + -0.00)


| Location | 13,889,156 – 13,889,270 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.32 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -20.88 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13889156 114 + 20766785 UCUGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGCCCCAGCUUUUCUUCUCCUCCUUUCCCCUUUC---UUCAUUCCCCUCUCUCCUC .......((..(((((((.((((..(((((((..(.....)..)).))))).)))).))))))).))............................---................... ( -21.30) >DroPse_CAF1 171660 100 + 1 UCGGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGUCUCAUCUCGCAU---CCCCAUU--------C---CCCC---CCCUCUCCUCUC ..((((.(((.(((((((.((((..(((((((..(.....)..)).))))).)))).)))))(........))).)---))...))--------)---)...---............ ( -22.10) >DroSec_CAF1 174995 114 + 1 UCUGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGCCCCAGCUUUUCUUCUCCUCCUUUCGCCUUUC---UCCACUUCCUUCUCUCCUC ...(((.(((((((((((.((((..(((((((..(.....)..)).))))).)))).)))))))....((..................)).....---.....)))))))....... ( -23.17) >DroSim_CAF1 178037 114 + 1 UCUGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGCCCCAGCUUUUCUUCUCCUCCUUUCGCCUUUC---UCCACUCCCUUCUCUCCUC ...(((.(((.(((((((.((((..(((((((..(.....)..)).))))).)))).)))))))....((..................)).....---.....))).)))....... ( -22.87) >DroEre_CAF1 175845 107 + 1 UCUGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGCCCCAGCUUUUCUUCUCGUCCUUUGCCCCU----------UCCCCUCGCUGCUC ...((..(((((((((((.((((..(((((((..(.....)..)).))))).)))).)))))))....((.................))...)----------)))..))....... ( -23.03) >DroYak_CAF1 180589 117 + 1 UCUGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGCCCCAGCUUUUCUUCUCGUCCUCUCUCCUUUCCAUUCCAUUCCCUGCGCUUCUC .......(((((((((((.((((..(((((((..(.....)..)).))))).)))).)))))((....))........................................)))))). ( -23.00) >consensus UCUGAACGGAAGCGGAUAUUUUCAACGCGCCAUUGUUAUUUUUUGAGUGCGGGAAAUUGUUCGCCCCAGCUUUUCUUCUCCUCCUUUCCCCUUUC___UCCA_UCCCCUCUCUCCUC .......((..(((((((.((((..(((((((..(.....)..)).))))).)))).))))))).)).................................................. (-20.88 = -20.60 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:40 2006