
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,888,035 – 13,888,135 |
| Length | 100 |
| Max. P | 0.975590 |

| Location | 13,888,035 – 13,888,135 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -28.30 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
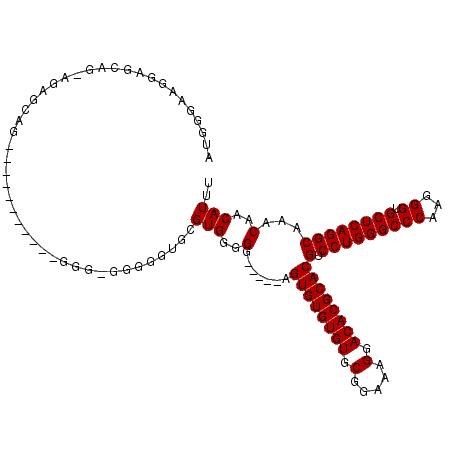
>2R_DroMel_CAF1 13888035 100 + 20766785 AUGGGAAGGAGCAGUAGAGCAG-------------------GGGCGUGGGGGGGUUGUGUGUGUGUGCGGGAAGGACACGCACGUCCUGGGCCCAAGGGUGCUCAGGCAAACAACAUUU ..........((....(((((.-------------------((((.(((((......(((((((((.(.....).)))))))))))))).)))).....)))))..))........... ( -35.70) >DroSec_CAF1 173897 113 + 1 AUGGAAAGGAGCAG-GGAGCAGGAGGGAGGGGGGGGGGGGGGUGCGUGAGGG-----AGUGUGUGUACGGAAAGGACACGCACGUCCUGGGCCCAAGGGUGCUCAGGCAAACAACAUUU ..........((..-...))................(..(..(((.((((((-----(((((((((.(.....).)))))))).)))...(((....))).)))).)))..)..).... ( -34.70) >DroSim_CAF1 177691 99 + 1 AUGGGAAG---CAG-UAAGCAG-----------GGGAGGGGGUGCGUGGGGG-----AGUGUGUGUGCGGAAAGGACACGCACGUCCUGGGCCCAAGGGUGCUCAGGCAAACAACAUUU .......(---(..-...))..-----------......(..(((.((((((-----(((((((((.(.....).)))))))).)))...(((....))).)))).)))..)....... ( -34.30) >consensus AUGGGAAGGAGCAG_AGAGCAG___________GGG_GGGGGUGCGUGGGGG_____AGUGUGUGUGCGGAAAGGACACGCACGUCCUGGGCCCAAGGGUGCUCAGGCAAACAACAUUU .............................................(((..(.......((((((((.(.....).))))))))(.(((((((((...)).))))))))...)..))).. (-28.30 = -28.30 + 0.00)

| Location | 13,888,035 – 13,888,135 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -22.61 |
| Consensus MFE | -18.47 |
| Energy contribution | -18.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
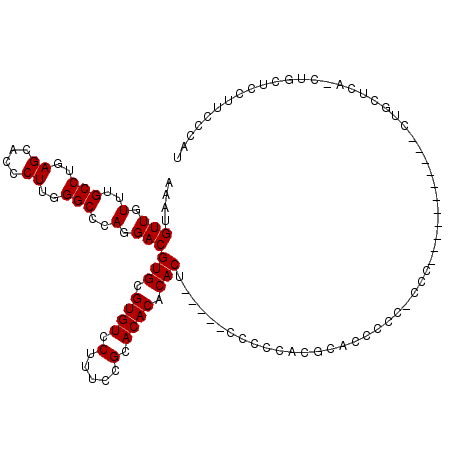
>2R_DroMel_CAF1 13888035 100 - 20766785 AAAUGUUGUUUGCCUGAGCACCCUUGGGCCCAGGACGUGCGUGUCCUUCCCGCACACACACACAACCCCCCCACGCCC-------------------CUGCUCUACUGCUCCUUCCCAU ....(((((.((((((.((.((...)))).))))..(((((.(.....).)))))...)).)))))............-------------------..((......)).......... ( -22.60) >DroSec_CAF1 173897 113 - 1 AAAUGUUGUUUGCCUGAGCACCCUUGGGCCCAGGACGUGCGUGUCCUUUCCGUACACACACU-----CCCUCACGCACCCCCCCCCCCCCCUCCCUCCUGCUCC-CUGCUCCUUUCCAU ...............(((((.....(((..(((((.(((((((........((....))...-----....))))))).................)))))..))-))))))........ ( -23.53) >DroSim_CAF1 177691 99 - 1 AAAUGUUGUUUGCCUGAGCACCCUUGGGCCCAGGACGUGCGUGUCCUUUCCGCACACACACU-----CCCCCACGCACCCCCUCCC-----------CUGCUUA-CUG---CUUCCCAU ...........((.((((((.....(((...(((..(((((((...................-----....)))))))..))).))-----------)))))))-..)---)....... ( -21.70) >consensus AAAUGUUGUUUGCCUGAGCACCCUUGGGCCCAGGACGUGCGUGUCCUUUCCGCACACACACU_____CCCCCACGCACCCCC_CCC___________CUGCUCA_CUGCUCCUUCCCAU ....(((.(..(((..((....))..)))..).)))(((.((((.(.....).)))).))).......................................................... (-18.47 = -18.47 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:38 2006