| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,870,982 – 13,871,080 |
| Length | 98 |
| Max. P | 0.837450 |

| Location | 13,870,982 – 13,871,080 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
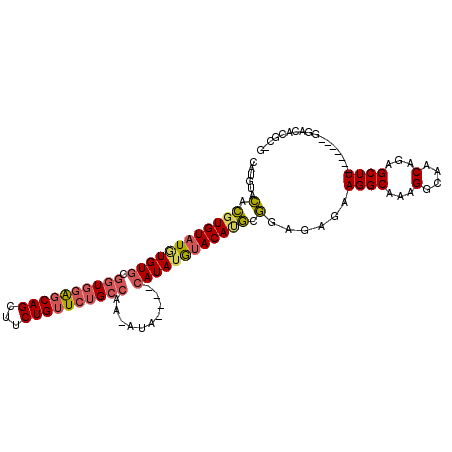
>2R_DroMel_CAF1 13870982 98 - 20766785 CAUGUACACGUGUAUGUGUGCGGUGGAGCAGCUUCUGUUCUGCCAA-AUA----CAUAUGUACAGGCGGAGAGAAGGCAAAGGCAACAGAGCUU-------GGACACGC-G ..((((((....(((((((..((..((((((...))))))..))..-)))----)))))))))).(((......((((...(....)...))))-------.....)))-. ( -34.70) >DroSec_CAF1 170935 98 - 1 CAUGUACUCGUGUAUGUGUGCGGUGGAGCAGCUUCUGUUCUGCCAA-AUA----CAUAUGUACAUGCGGAGAGAAGGCAAAGGCAACAGAGCUU-------GGACACGC-G .((((((....((((((((..((..((((((...))))))..))..-)))----)))))))))))(((......((((...(....)...))))-------.....)))-. ( -35.50) >DroSim_CAF1 174792 98 - 1 CAUGUACACGUGUAUGUGUGCGGUGGAGCAGCUUCUGUUCUGCCAA-AUA----CAUAUGUACAUGCGGAGAGAAGGCAAAGGCAACAGAGCUU-------GGACACGC-G .(((((((....(((((((..((..((((((...))))))..))..-)))----)))))))))))(((......((((...(....)...))))-------.....)))-. ( -35.70) >DroEre_CAF1 171807 101 - 1 CAUGUACACGUGUAUGUGUGCGGUGGAGCAGCUUCUGUUCUGCAAA-AUACAUACAUAUGUACAUGCGGAGAGAAGGCA-AGGCAACAGAGCUU-------GGACACGC-G .(((((((..(((((((((...(..((((((...))))))..)...-)))))))))..)))))))(((......((((.-.(....)...))))-------.....)))-. ( -41.70) >DroWil_CAF1 208191 103 - 1 --UGUAUAUGUGUGUUUGUAUG-UUUGUCAGCUUCUGUUCUGGCAAAAGA----CAUACAGACACACACACAC-AGGCCAGGCAAACAGAGCUUAAUGCUUGGAGUUAUUU --(((.(.((((((((((((((-((((((((........))))))...))----)))))))))))))).).))-)..(((((((............)))))))........ ( -37.80) >DroYak_CAF1 176597 98 - 1 CAUGUACACGUGUAUAUGUGCGGUGGAGCAGCUUCUGUUCUGCCAA-AUA----CAUAUGUACAUGCGGAGAGAAGGCAAAGGCAACAGAGCUU-------GGACACGC-G .(((((((..(((((......((..((((((...))))))..))..-)))----))..)))))))(((......((((...(....)...))))-------.....)))-. ( -34.10) >consensus CAUGUACACGUGUAUGUGUGCGGUGGAGCAGCUUCUGUUCUGCCAA_AUA____CAUAUGUACAUGCGGAGAGAAGGCAAAGGCAACAGAGCUU_______GGACACGC_G ......(.((((((((((((.((((((((((...))))))))))..........)))))))))))).)......((((...(....)...))))................. (-21.23 = -21.68 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:37 2006