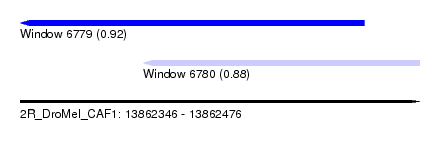
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,862,346 – 13,862,476 |
| Length | 130 |
| Max. P | 0.917767 |

| Location | 13,862,346 – 13,862,458 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -14.80 |
| Energy contribution | -16.36 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13862346 112 - 20766785 UUUACAGAUGCAGAUACAGAUA------CAGAUCCAGAU--AGAUAUGGAGUACACUUUGUAUUUGUUAUUCUGUGAAAGCAAAUAAAAUACAUUUUCUAUCUGUCUAUCUCCAUUCGCA ....(..(((.(((((((((((------((((....(((--(((((..(((....)))..))))))))..)))))(((((.............))))).))))))..)))).)))..).. ( -25.92) >DroSec_CAF1 162294 120 - 1 UUUACCAAUGCAGAUACAGAUGCAGAUGCAGAUACAGAUACAGAUAUGGAGUACGAUUUGUAUUUGUUAUUCUGUGAAAGCAAAUAAAAUACAUUUUCUAUCUGUCUAUCUCCAUUCGCA ........(((..((..(((((((((((((((.((((((((((((.((.....))))))))))))))...)))))(((((.............))))).))))))..))))..))..))) ( -29.52) >DroSim_CAF1 167595 114 - 1 UUUACCAAUGCAGAUACAGAUG------CAGAUACAGAUACAGAUAUGGAGUACGAUUUGUAUUUGUUAUUCUGUGAAAGCAAAUAAAAUACAUUUUCUAUCUGUCUAUCUCCAUUCGCA ......((((.(((((((((((------((((.((((((((((((.((.....))))))))))))))...)))))(((((.............))))).))))))..)))).)))).... ( -28.12) >DroEre_CAF1 163237 104 - 1 ----------CCGAUACAGAUA------CAGAUACAGAUACGGAUAUGGAGUACACUUUGUAUUUGUUAUGCUGUGAAAGCAAAUAAAAUACAUUUUCUAUCUGUCUAUCUGAAUUCGCA ----------.(((..((((((------((((((.(((.(((((((..(((....)))..)))))))..((((.....))))............))).)))))))..)))))...))).. ( -28.00) >DroYak_CAF1 167996 90 - 1 ------------------------------GAUACAGUUACGGAUAUGGAGUACACUUUGUAUUUGCUAUUCUGUGAAAGCAAAUAAAAUACAUUUUCUAUCUGUCUAUCUCAUUUCGCA ------------------------------((((.....(((((((.((((.........((((((((.(((...)))))))))))........))))))))))).)))).......... ( -16.13) >consensus UUUAC__AUGCAGAUACAGAUA______CAGAUACAGAUACAGAUAUGGAGUACACUUUGUAUUUGUUAUUCUGUGAAAGCAAAUAAAAUACAUUUUCUAUCUGUCUAUCUCCAUUCGCA .............................(((((((((((((((((..(((....)))..))))))(((((.(((....))))))))...........)))))))..))))......... (-14.80 = -16.36 + 1.56)

| Location | 13,862,386 – 13,862,476 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -21.18 |
| Consensus MFE | -12.41 |
| Energy contribution | -13.72 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13862386 90 - 20766785 GAUACAGAUACAGACACAUUUACAGAUGCAGAUACAGAUA------CAGAUCCAGAU--AGAUAUGGAGUACACUUUGUAUUUGUUAUUCUGUGAAAG ..................((((((((.(((((((((((..------....((((...--.....))))......)))))))))))...)))))))).. ( -25.30) >DroSec_CAF1 162334 98 - 1 GAUACAGAUACAGAUACAUUUACCAAUGCAGAUACAGAUGCAGAUGCAGAUACAGAUACAGAUAUGGAGUACGAUUUGUAUUUGUUAUUCUGUGAAAG ..........................((((........))))...(((((.((((((((((((.((.....))))))))))))))...)))))..... ( -22.10) >DroSim_CAF1 167635 86 - 1 GAUA------CAGAUACAUUUACCAAUGCAGAUACAGAUG------CAGAUACAGAUACAGAUAUGGAGUACGAUUUGUAUUUGUUAUUCUGUGAAAG ..((------((((............((((........))------))...((((((((((((.((.....))))))))))))))...)))))).... ( -21.20) >DroEre_CAF1 163277 80 - 1 GAUACAGAUACCGAUA------------CCGAUACAGAUA------CAGAUACAGAUACGGAUAUGGAGUACACUUUGUAUUUGUUAUGCUGUGAAAG ................------------............------....(((((..(((((((..(((....)))..)))))))....))))).... ( -16.10) >consensus GAUACAGAUACAGAUACAUUUACCAAUGCAGAUACAGAUA______CAGAUACAGAUACAGAUAUGGAGUACAAUUUGUAUUUGUUAUUCUGUGAAAG ..................................................((((((.(((((((..(((....)))..)))))))...)))))).... (-12.41 = -13.72 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:36 2006