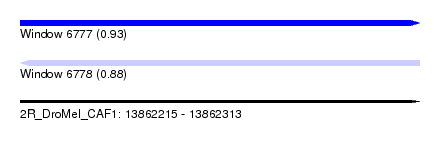
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,862,215 – 13,862,313 |
| Length | 98 |
| Max. P | 0.925231 |

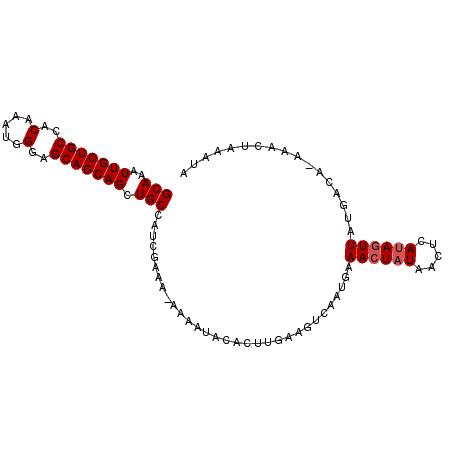
| Location | 13,862,215 – 13,862,313 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.26 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -19.05 |
| Energy contribution | -18.29 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
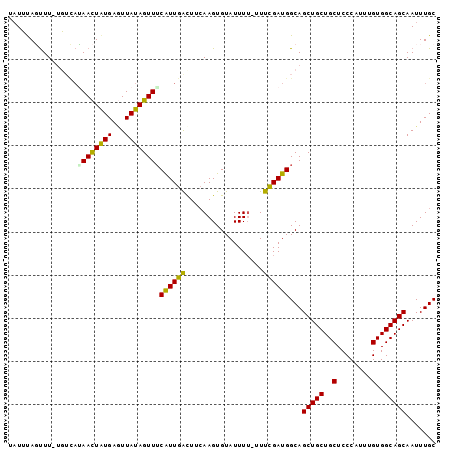
>2R_DroMel_CAF1 13862215 98 + 20766785 UAUUUAGUUU-UGUCAUAACUAUGAGUUAUAGUUUCAUUGAUUUCAAGUGUAUUUUUUUUCGAUGGCAGCUGCUGCUCCCAUUUGUGGCAGCAAUUUGC .........(-((((((((....((((..((((((((((((..................))))))).)))))..))))....)))))))))........ ( -22.07) >DroSec_CAF1 162164 97 + 1 UAUUUAGUUU-UGUCAUAACUAUGAGUUAUAGUUUCAUUGGCUUCAAGUGUAUUUU-UUUCGAUGGCAGCUGCUGCUCCCAUUUGUGGCAGCAAUUUGC ..........-(((((((((((((...))))))).((((.......))))......-.....))))))(((((..(........)..)))))....... ( -22.00) >DroSim_CAF1 167464 98 + 1 AAUUUAGUUU-UGUCAUAACUGUGAGUUAUAGUUACAUUGACUUCAAGUGUAUUUUUUUUCGAUGGCAGCUGCUGCUCCCAUUUGUGGCAGCAAUUUGC ..........-(((((((((((((...)))))))(((((.......)))))...........))))))(((((..(........)..)))))....... ( -24.50) >DroEre_CAF1 163109 95 + 1 U-UGUAUCUUUAUGGUGAAUUAUGAUUUAUAGUUUCGUUGAUUUCAAGUGUAUUU---UUCGAUGGCAGCUGCUGCUCCCAUUUGUGGCAGCAAUUUGC .-............(..((((.............(((((((..............---.)))))))..(((((..(........)..)))))))))..) ( -20.16) >DroYak_CAF1 167866 97 + 1 UAUGCAUUUU-UUGUAAAACUAUGAAUUAUAGUUUCGUUGGCUUGAAGUGUAUUUU-UUUUGAUGGCAGCUGCUGCUCCCAUUUGUGGCAGCAAUUUGC .(((((((((-..((.((((((((...)))))))).....))..)))))))))...-...........(((((..(........)..)))))....... ( -25.60) >consensus UAUUUAGUUU_UGUCAUAACUAUGAGUUAUAGUUUCAUUGACUUCAAGUGUAUUUU_UUUCGAUGGCAGCUGCUGCUCCCAUUUGUGGCAGCAAUUUGC ................((((((((...))))))))((((((..................))))))...(((((..(........)..)))))....... (-19.05 = -18.29 + -0.76)

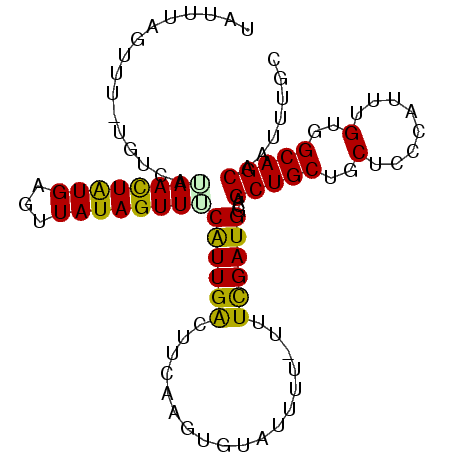
| Location | 13,862,215 – 13,862,313 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 87.26 |
| Mean single sequence MFE | -16.40 |
| Consensus MFE | -13.50 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13862215 98 - 20766785 GCAAAUUGCUGCCACAAAUGGGAGCAGCAGCUGCCAUCGAAAAAAAAUACACUUGAAAUCAAUGAAACUAUAACUCAUAGUUAUGACA-AAACUAAAUA (((..(((((((..(.....)..))))))).)))...............((.(((....)))))....((((((.....))))))...-.......... ( -16.20) >DroSec_CAF1 162164 97 - 1 GCAAAUUGCUGCCACAAAUGGGAGCAGCAGCUGCCAUCGAAA-AAAAUACACUUGAAGCCAAUGAAACUAUAACUCAUAGUUAUGACA-AAACUAAAUA (((..(((((((..(.....)..))))))).)))........-.........................((((((.....))))))...-.......... ( -15.70) >DroSim_CAF1 167464 98 - 1 GCAAAUUGCUGCCACAAAUGGGAGCAGCAGCUGCCAUCGAAAAAAAAUACACUUGAAGUCAAUGUAACUAUAACUCACAGUUAUGACA-AAACUAAAUU (((..(((((((..(.....)..))))))).))).............((((.(((....)))))))..((((((.....))))))...-.......... ( -18.70) >DroEre_CAF1 163109 95 - 1 GCAAAUUGCUGCCACAAAUGGGAGCAGCAGCUGCCAUCGAA---AAAUACACUUGAAAUCAACGAAACUAUAAAUCAUAAUUCACCAUAAAGAUACA-A (((..(((((((..(.....)..))))))).))).......---.....................................................-. ( -12.70) >DroYak_CAF1 167866 97 - 1 GCAAAUUGCUGCCACAAAUGGGAGCAGCAGCUGCCAUCAAAA-AAAAUACACUUCAAGCCAACGAAACUAUAAUUCAUAGUUUUACAA-AAAAUGCAUA (((..(((((((..(.....)..)))))))............-....................((((((((.....))))))))....-....)))... ( -18.70) >consensus GCAAAUUGCUGCCACAAAUGGGAGCAGCAGCUGCCAUCGAAA_AAAAUACACUUGAAGUCAAUGAAACUAUAACUCAUAGUUAUGACA_AAACUAAAUA (((..(((((((..(.....)..))))))).)))...............................((((((.....))))))................. (-13.50 = -13.90 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:34 2006