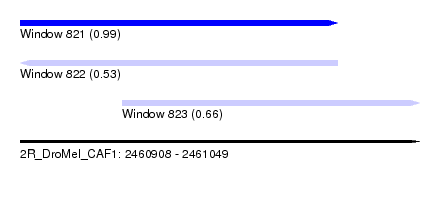
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,460,908 – 2,461,049 |
| Length | 141 |
| Max. P | 0.990189 |

| Location | 2,460,908 – 2,461,020 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.73 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -31.50 |
| Energy contribution | -31.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
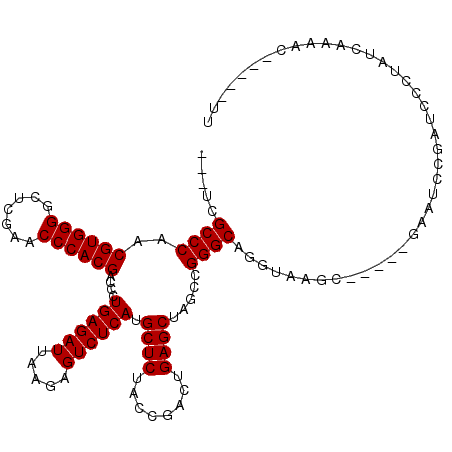
>2R_DroMel_CAF1 2460908 112 + 20766785 --ACUAUUAAAAUUUGAAUAUUUACUACA-AAAUAUUGG---UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAG --............(.(((((((......-))))))).)---..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).. ( -33.50) >DroPse_CAF1 32327 110 + 1 --CCUAUUGCUUC---AAAAGAUAGACCAAAAAUAAAUG---UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAU --.(((((.....---....)))))..............---..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).. ( -33.10) >DroAna_CAF1 19152 115 + 1 GAAUGACAAAAAAAAGAAACGC--GACCA-AAAUAAUGAAAUGCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAU ....................((--(..((-......))...)))((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).. ( -32.30) >DroPer_CAF1 23364 110 + 1 --CCUAUUGCUUC---AAAAGAUAGACCAAAAAUAAAUG---UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAU --.(((((.....---....)))))..............---..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).. ( -33.10) >consensus __ACUAUUAAAAC___AAAAGAUAGACCA_AAAUAAAGG___UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAU ............................................((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).. (-31.50 = -31.50 + 0.00)

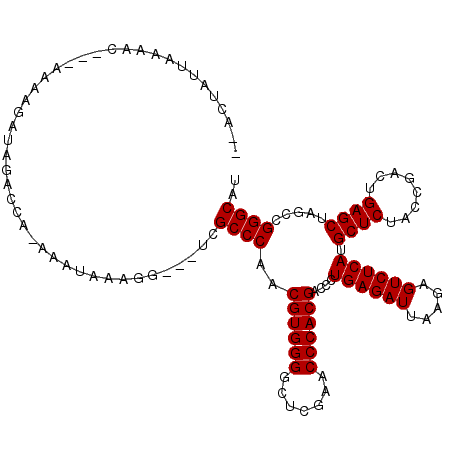
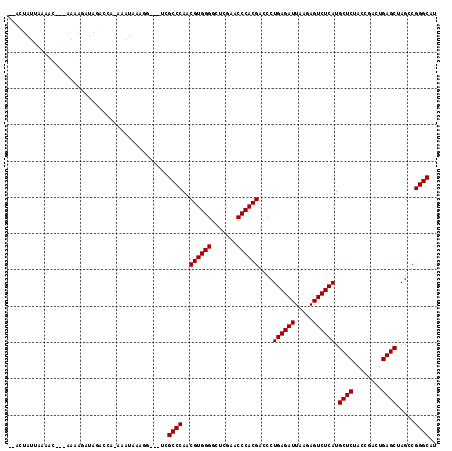
| Location | 2,460,908 – 2,461,020 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.73 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -32.88 |
| Energy contribution | -32.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2460908 112 - 20766785 CUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGA---CCAAUAUUU-UGUAGUAAAUAUUCAAAUUUUAAUAGU-- .((((((((..((((........))))....((((((.......))))))(((((.......))))))))))))).---..(((((((-......)))))))..............-- ( -34.30) >DroPse_CAF1 32327 110 - 1 AUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGA---CAUUUAUUUUUGGUCUAUCUUUU---GAAGCAAUAGG-- ..(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))((---(...........)))........---...........-- ( -33.60) >DroAna_CAF1 19152 115 - 1 AUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGCAUUUCAUUAUUU-UGGUC--GCGUUUCUUUUUUUUGUCAUUC ..(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))((...(((......-)))..--)).................... ( -35.10) >DroPer_CAF1 23364 110 - 1 AUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGA---CAUUUAUUUUUGGUCUAUCUUUU---GAAGCAAUAGG-- ..(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))((---(...........)))........---...........-- ( -33.60) >consensus AUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGA___CAAUUAUUU_UGGUCUAUCUUUU___GAAGCAAUAGG__ .((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))........................................... (-32.88 = -32.88 + 0.00)

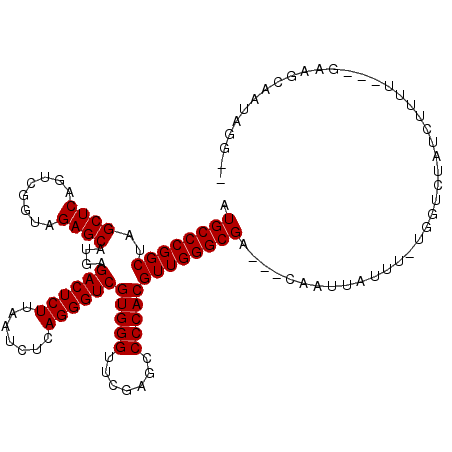
| Location | 2,460,944 – 2,461,049 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -31.50 |
| Energy contribution | -31.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2460944 105 + 20766785 ---UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAAU-----GAAUUUGCUGUCUGCCAAAAC-----UC ---..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).((((.((-----.........)).)))).....-----.. ( -34.20) >DroPse_CAF1 32361 115 + 1 ---UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGGGCCAAUCGAAUGCGAUCCGUAUCCGGCCAACACCU ---..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).(((.((((....(.(((((...))))).))))).)))... ( -41.60) >DroSim_CAF1 23173 105 + 1 ---UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAACC-----GAAUUUGGUGUCUGUGAAAAG-----UU ---..((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))((..(.((-----(....))))..))........-----.. ( -34.10) >DroYak_CAF1 21839 105 + 1 ---UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAAC-----GAAUCCGCUGACUGUCAAAAC-----UC ---..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).(((...(-----(....))...)))........-----.. ( -33.00) >DroAna_CAF1 19188 106 + 1 AAUGCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGGGC-----GUAUCCGAUCGGUAUCAGAAC------- .(((((((((.((((((.......)))))).(((((((((.....)))))).((((........)))).....)))....)))))-----))))((....)).........------- ( -42.20) >DroPer_CAF1 23398 115 + 1 ---UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGGGCCAAUCGAAUGCGAUCCGUAUCCGGCCAACACCU ---..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).(((.((((....(.(((((...))))).))))).)))... ( -41.60) >consensus ___UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAGC_____GAAUCCGAUCCCUAUCAAAAC_____UU .....((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))......................................... (-31.50 = -31.50 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:14 2006