
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,859,569 – 13,859,669 |
| Length | 100 |
| Max. P | 0.981096 |

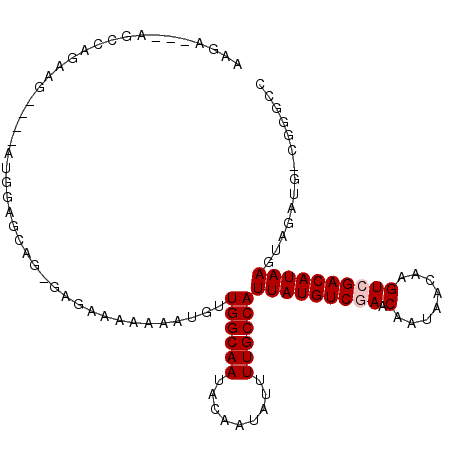
| Location | 13,859,569 – 13,859,669 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.76 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
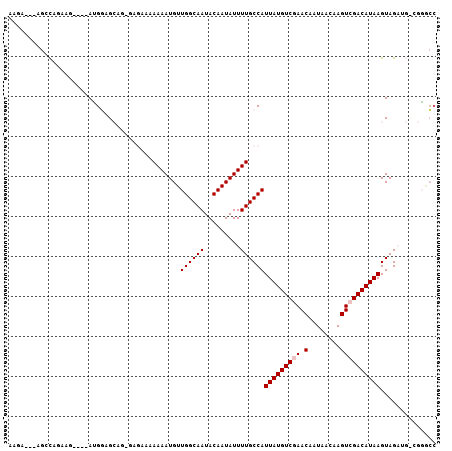
>2R_DroMel_CAF1 13859569 100 + 20766785 AAGA---AGCCAGAAG----ACGGAGCAGGGAGAAAAAAAUGUUGGCAAUACAAUAUUUUGCCAUUAUGUCGAACAAUAACAAGUCGACAUAAGUAGAUGUCGGGCC ....---.(((....(----(((..((..((.....(((((((((......))))))))).)).(((((((((.(........))))))))))))...)))).))). ( -28.20) >DroVir_CAF1 198177 87 + 1 ----------CAAGCC----AAGCAGC------ACAAAAAUGUUGGCAAUACAAUAUUUUGCCAUUAUGUCGAACAAUAACAAGUCGACAUAAGUAGAUGCCGUCGC ----------...((.----..((.((------((........((((((.........))))))(((((((((.(........))))))))))...).))).)).)) ( -20.00) >DroPse_CAF1 156084 101 + 1 AAGAAGAGGAUAGAAGGAGCAUGGAGAAGAGAG-AAAAAAUGUUGGCAAUACAAUAUUUUGCCAUUAUGUCGAACAAUAACAAGUAGACAUAAGUAGU-----GGCC .................................-..(((((((((......)))))))))(((((((((((..((........)).))))))....))-----))). ( -18.50) >DroGri_CAF1 188391 94 + 1 GCCA---AGCCAAGCC----AAGCAGC------ACAAAAAUGUUGGCAAUACAAUAUUUUGCCAUUAUGUCGAACAAUAACAAGUCGACAUAAGUAGAUGCCGUACA ....---.........----..((.((------((........((((((.........))))))(((((((((.(........))))))))))...).))).))... ( -20.30) >DroEre_CAF1 160480 99 + 1 AAGA---AGCCAGAAG----AUGGAGCAGGGAGAA-AAAAUGUUGGCAAUACAAUAUUUUGCCAUUAUGUCGAACAAUAACAAGUCGACAUAAGUAGAUGUCGGGCC ....---.(((....(----(((..((..((....-(((((((((......))))))))).)).(((((((((.(........))))))))))))...)))).))). ( -26.40) >DroPer_CAF1 156696 101 + 1 AAGAAGAGGAUAGAAGGAGCAUGGAGAAGAGAG-AAAAAAUGUUGGCAAUACAAUAUUUUGCCAUUAUGUCGAACAAUAACAAGUAGACAUAAGUAGC-----GACC ...............(..((.(((.........-..(((((((((......))))))))).)))(((((((..((........)).)))))))...))-----..). ( -18.10) >consensus AAGA___AGCCAGAAG____AUGGAGCAG_GAGAAAAAAAUGUUGGCAAUACAAUAUUUUGCCAUUAUGUCGAACAAUAACAAGUCGACAUAAGUAGAUG_CGGGCC ...........................................((((((.........))))))(((((((((.(........)))))))))).............. (-14.57 = -14.90 + 0.33)

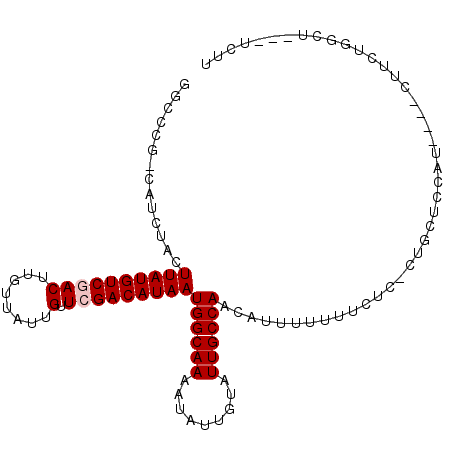
| Location | 13,859,569 – 13,859,669 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.76 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -14.17 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
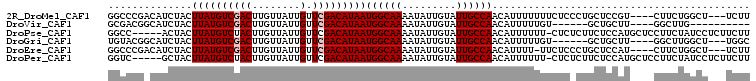
>2R_DroMel_CAF1 13859569 100 - 20766785 GGCCCGACAUCUACUUAUGUCGACUUGUUAUUGUUCGACAUAAUGGCAAAAUAUUGUAUUGCCAACAUUUUUUUCUCCCUGCUCCGU----CUUCUGGCU---UCUU ((((.(((......((((((((((........).)))))))))((((((.........)))))).....................))----)....))))---.... ( -23.20) >DroVir_CAF1 198177 87 - 1 GCGACGGCAUCUACUUAUGUCGACUUGUUAUUGUUCGACAUAAUGGCAAAAUAUUGUAUUGCCAACAUUUUUGU------GCUGCUU----GGCUUG---------- .(((((((((....((((((((((........).)))))))))((((((.........))))))........))------)))).))----).....---------- ( -24.30) >DroPse_CAF1 156084 101 - 1 GGCC-----ACUACUUAUGUCUACUUGUUAUUGUUCGACAUAAUGGCAAAAUAUUGUAUUGCCAACAUUUUUU-CUCUCUUCUCCAUGCUCCUUCUAUCCUCUUCUU (((.-----.....(((((((.((........))..)))))))((((((.........)))))).........-.............)))................. ( -13.90) >DroGri_CAF1 188391 94 - 1 UGUACGGCAUCUACUUAUGUCGACUUGUUAUUGUUCGACAUAAUGGCAAAAUAUUGUAUUGCCAACAUUUUUGU------GCUGCUU----GGCUUGGCU---UGGC .....(((((....((((((((((........).)))))))))((((((.........))))))........))------)))(((.----(((...)))---.))) ( -24.40) >DroEre_CAF1 160480 99 - 1 GGCCCGACAUCUACUUAUGUCGACUUGUUAUUGUUCGACAUAAUGGCAAAAUAUUGUAUUGCCAACAUUUU-UUCUCCCUGCUCCAU----CUUCUGGCU---UCUU ((((.((.......((((((((((........).)))))))))((((((.........)))))).......-...............----..)).))))---.... ( -22.10) >DroPer_CAF1 156696 101 - 1 GGUC-----GCUACUUAUGUCUACUUGUUAUUGUUCGACAUAAUGGCAAAAUAUUGUAUUGCCAACAUUUUUU-CUCUCUUCUCCAUGCUCCUUCUAUCCUCUUCUU ((..-----((...(((((((.((........))..)))))))((((((.........)))))).........-.............)).))............... ( -15.40) >consensus GGCCCG_CAUCUACUUAUGUCGACUUGUUAUUGUUCGACAUAAUGGCAAAAUAUUGUAUUGCCAACAUUUUUUUCUC_CUGCUCCAU____CUUCUGGCU___UCUU ..............((((((((((........).)))))))))((((((.........))))))........................................... (-14.17 = -14.50 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:32 2006