
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,850,753 – 13,850,846 |
| Length | 93 |
| Max. P | 0.540738 |

| Location | 13,850,753 – 13,850,846 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -20.93 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540738 |
| Prediction | RNA |
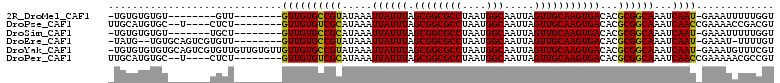
Download alignment: ClustalW | MAF
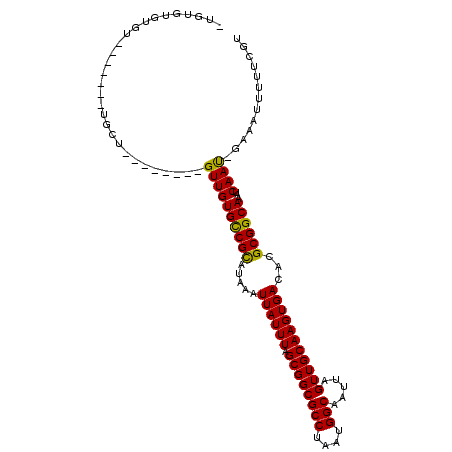
>2R_DroMel_CAF1 13850753 93 + 20766785 -UGUGUGUGU--------GUU--------GUUGUGCCGUAUAAAUUAUUUAGCGGCGCCUAAUGGCAAUUAGUUGCAAGUGACACGCGGCAAAUCAAU-GAAAUUUUUGGU -(((.(((((--------((.--------.(((((((((............)))))))......((((....)))).))..)))))))))).(((((.-.......))))) ( -26.80) >DroPse_CAF1 149383 97 + 1 UUGCAUGUGC--U----CUCU--------GUUGUGUCGCAUAAAUUAUUUAGCGGCGCCUAAUGGCAAUUAGUUGCAAGUGACACGCGGCAAAUCAACCGAAAACCGACGU ..((....))--.----.((.--------((((((((((.....((((((.((((((((....))).....)))))))))))...))))))...)))).)).......... ( -23.00) >DroSim_CAF1 156189 94 + 1 -UGUGUGUGU-------UGCU--------GUUGUGCCGCAUAAAUUAUUUAGCGGCGCCUAAUGGCAAUUAGUUGCAAGUGACACGCGGCAAAUCAAU-GAAAUUUUUGGU -(((((((((-------..((--------...(((((((.(((.....))))))))))......((((....)))).))..)))))).))).(((((.-.......))))) ( -31.50) >DroEre_CAF1 151957 98 + 1 -UAUG--UGUGCAGUCGUGUU--------GUUGUGCCGUAUAAAUUAUUUAGCGGCGCCUAAUGGCAAUUAGUUGCAAGUGACACGCGGCAAAUCAAU-GAAAU-UUUUGU -..((--..(((.(.(((((.--------.(((((((((............)))))))......((((....)))).))..)))))).)))...))..-.....-...... ( -27.30) >DroYak_CAF1 156299 109 + 1 -UGUGUGUGUGCAGUCGUGUUGUUGUGUUGUUGUGCCGUAUAAAUUAUUUAGCGGCGCCUAAUGGCAAUUAGUUGCAAGUGACACGCGGCAAAUCAAU-GAAAUGUUUCGU -.((((((.(((((.(..(((((..(......(((((((............)))))))...)..)))))..))))))....)))))).........((-(((....))))) ( -29.20) >DroPer_CAF1 149979 97 + 1 UUGCAUGUGC--U----CUCU--------GUUGUGUCGCAUAAAUUAUUUAGCGGCGCCUAAUGGCAAUUAGUUGCAAGUGACACGCGGCAAAUCAACCGAAAAACGCCGU .(((((.(((--.----..((--------(....(((((.(((.....))))))))(((....)))...)))..))).))).)).(((((................))))) ( -24.79) >consensus _UGUGUGUGU_______UGCU________GUUGUGCCGCAUAAAUUAUUUAGCGGCGCCUAAUGGCAAUUAGUUGCAAGUGACACGCGGCAAAUCAAU_GAAAUUUUUCGU .............................((((((((((.....((((((.((((((((....))).....)))))))))))...))))))...))))............. (-20.93 = -20.23 + -0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:26 2006