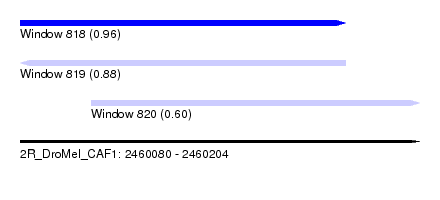
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,460,080 – 2,460,204 |
| Length | 124 |
| Max. P | 0.963660 |

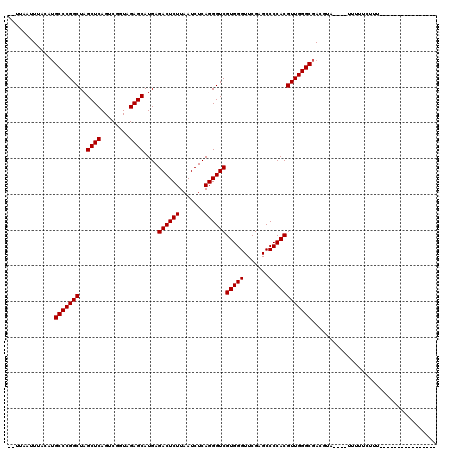
| Location | 2,460,080 – 2,460,181 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -31.42 |
| Energy contribution | -31.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2460080 101 + 20766785 ----------------ACUACAAAAUAU--UGGUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAACAAAA- ----------------............--.....((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))............- ( -31.50) >DroPse_CAF1 31926 110 + 1 CAGAAAAAUGAC-A--AAAGAAAAA----AACGUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUAAUUUA--- ............-.--.........----.((((.((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))))))......--- ( -33.30) >DroEre_CAF1 21227 90 + 1 -----------------------------UGCAUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAACGAAU- -----------------------------(((...((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))..))).......- ( -32.10) >DroYak_CAF1 20962 103 + 1 ----------------AGAGUAAAAUAUAUUGAUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGAAUUUGG- ----------------...................((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))............- ( -31.50) >DroAna_CAF1 18608 110 + 1 ---------GGCCAAAAAUAAAAAAUUG-UAAGUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUUGAAAUUAGG ---------(((.....((((....)))-).....)))...((((((.......))))))..(((((..((((.(((((...((((........)))).....))))).))))..))))) ( -33.00) >DroPer_CAF1 22964 110 + 1 CAGAAAAAUGAC-A--AAAGAAAAA----AACGUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUAAUUUA--- ............-.--.........----.((((.((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))))))......--- ( -33.30) >consensus ________________AAAGAAAAA____UACGUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAAUUAA__ ...................................((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))............. (-31.42 = -31.42 + 0.00)

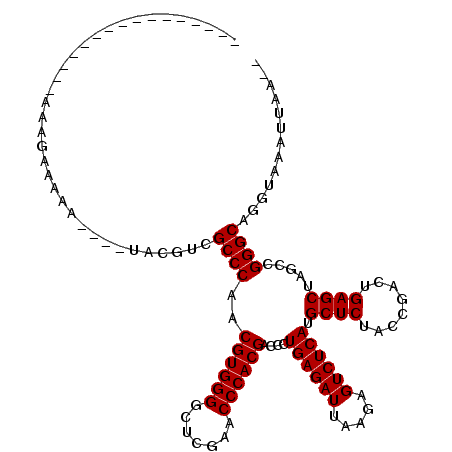
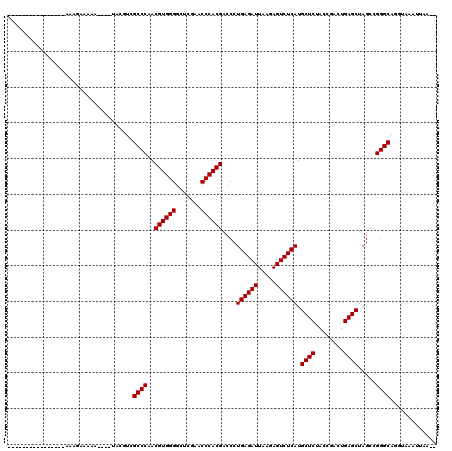
| Location | 2,460,080 – 2,460,181 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -32.73 |
| Energy contribution | -32.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2460080 101 - 20766785 -UUUUGUUUACCUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGACCA--AUAUUUUGUAGU---------------- -...........((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))....--............---------------- ( -32.80) >DroPse_CAF1 31926 110 - 1 ---UAAAUUACAUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGACGUU----UUUUUCUUU--U-GUCAUUUUUCUG ---..........(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))((((..----.........--)-)))......... ( -35.20) >DroEre_CAF1 21227 90 - 1 -AUUCGUUUACCUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAUGCA----------------------------- -...........((((((((..((((........))))....((((((.......))))))(((((.......))))))))))))).....----------------------------- ( -32.80) >DroYak_CAF1 20962 103 - 1 -CCAAAUUCACAUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAUCAAUAUAUUUUACUCU---------------- -...........((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))..................---------------- ( -33.00) >DroAna_CAF1 18608 110 - 1 CCUAAUUUCAACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGACUUA-CAAUUUUUUAUUUUUGGCC--------- .............(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))......-...................--------- ( -32.90) >DroPer_CAF1 22964 110 - 1 ---UAAAUUACAUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGACGUU----UUUUUCUUU--U-GUCAUUUUUCUG ---..........(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))((((..----.........--)-)))......... ( -35.20) >consensus __UUAAUUUACAUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGACGUA____UUUUUCUUU________________ .............(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))................................... (-32.73 = -32.73 + -0.00)

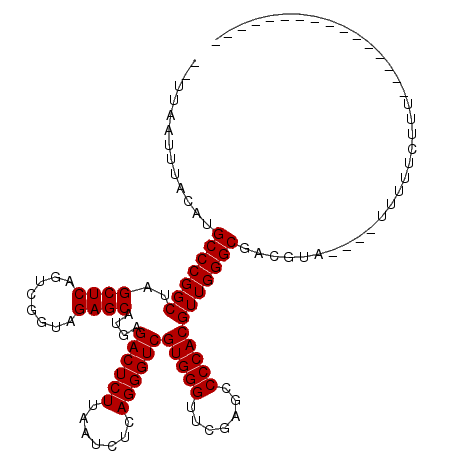
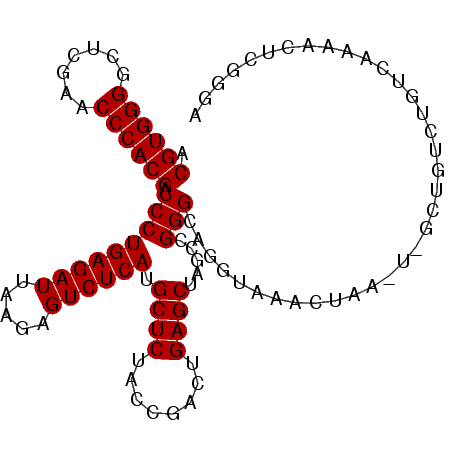
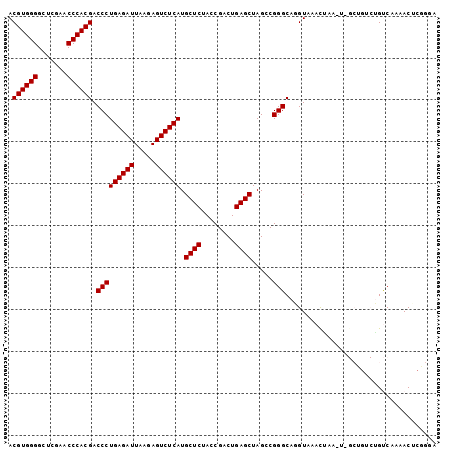
| Location | 2,460,102 – 2,460,204 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.74 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -26.90 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2460102 102 + 20766785 ACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAACAAAAUUGCCGUCUGUCAAAACUCGGGA .((((((.......)))))).(((((((((.....)))))).((((........))))..(.(((((.(((((......)))))))))).).......))). ( -37.40) >DroPse_CAF1 31959 86 + 1 ACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUAAUUUA----------GGCCAAAAC------ .((((((.......)))))).(((((((((.....)))))).((((........)))).....)))...........----------.........------ ( -26.90) >DroSim_CAF1 22401 102 + 1 ACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAACGGAUAUGCUGUCUGUCAAAACUCGGGA .((((((.......)))))).(((((((((.....)))))).((((........))))..(.((((((((((......))).))))))).).......))). ( -35.70) >DroEre_CAF1 21238 102 + 1 ACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAACGAAUUCGCUGUCUGUCAAAACUCGGGA .((((((.......)))))).(((((((((.....)))))).((((........))))..(.(((((((.....((....))))))))).).......))). ( -35.30) >DroYak_CAF1 20986 102 + 1 ACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGAAUUUGGUCGCUGGCUGUCAAAACGAUCUA .((((((.......))))))......(((((.(((((.....)))))...(((..(((((((.((.((........)).)))))))))))).....))))). ( -34.90) >DroPer_CAF1 22997 86 + 1 ACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUAAUUUA----------GGCCAAAAC------ .((((((.......)))))).(((((((((.....)))))).((((........)))).....)))...........----------.........------ ( -26.90) >consensus ACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAGGUAAACUAA_U_GCUGUCUGUCAAAACUCGGGA .((((((.......)))))).(((((((((.....)))))).((((........)))).....))).................................... (-26.90 = -26.90 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:12 2006