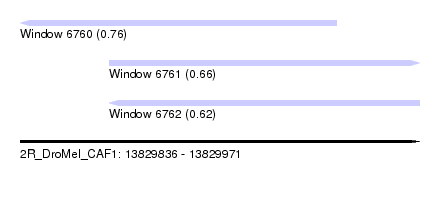
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,829,836 – 13,829,971 |
| Length | 135 |
| Max. P | 0.760070 |

| Location | 13,829,836 – 13,829,943 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -17.57 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13829836 107 - 20766785 GCCCACACAAACGAUGAAAACUAUGGCGUUGGGCGAAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAA-AAGGAAAACUAUAUGUGUGUACAAAAAAAAAAAAA-- (..((((((...........((((..((((((((............))))))))..))))............-.((.....))...))))))..).............-- ( -20.00) >DroEre_CAF1 131045 91 - 1 GCCCACACAAACG---------AUGGCGUUGGGCGAAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAAAAAGGAAAACUAUAUGUGUGUAUAUACA---------- ...((((((....---------((((((((((((............))))))))..(((........)))...........)))).))))))........---------- ( -16.40) >DroYak_CAF1 135104 100 - 1 GCCCACACAAACG---------AUGGCGUUGGGCAAAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAAAAAGGAAAACUAUAUGUGUGUAUAAAAAAAAAGAC-GA ...((((((....---------((((((((((((.(((....))).))))))))..(((........)))...........)))).))))))...............-.. ( -16.30) >consensus GCCCACACAAACG_________AUGGCGUUGGGCGAAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAAAAAGGAAAACUAUAUGUGUGUAUAAAAAAAAA_A____ ...((((((.................((((((((.(((....))).))))))))....................((.....))...)))))).................. (-16.00 = -16.00 + 0.00)

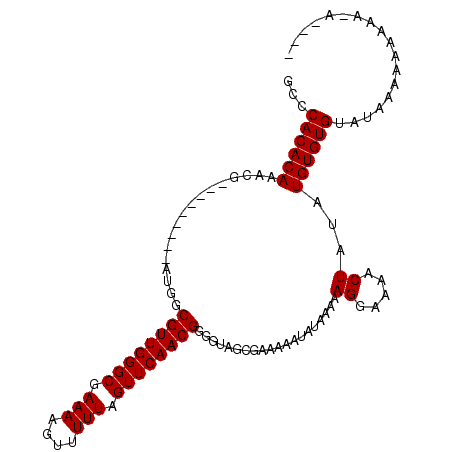
| Location | 13,829,866 – 13,829,971 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -18.08 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13829866 105 + 20766785 UCCUU-UUAUAUUUUUCGCUACCCCGUUGAACUAAAAACUUUU-CGCCCAACGCCAUAGUUUUCAUCGUUUGUGUGGGCGGGGUAAAUGCGUUUUUGAUGCUG---UUUU .....-...((((...((((((((((((........)))....-.(((((.(((....(.......)....))))))))))))))...))).....))))...---.... ( -25.20) >DroSec_CAF1 130420 104 + 1 UCCUU-UUAUAUUUUUCGCUACCCCGUUGAACUAAAAACUUUU-CGCCCAACGCCAUCGUUUUCAUCGUUUGUGUGGGCGGGGUAAAUGCGUUUUUGAUGCUG---UUU- .....-...((((...((((((((((((........)))....-.(((((.(((...((.......))...))))))))))))))...))).....))))...---...- ( -27.50) >DroSim_CAF1 135671 104 + 1 UCCUU-UUAUAUUUUUCGCUACCCCGUUGAACUAAAAACUUUU-CGCCCAACGCCAUCGUUUUCAUCGUUUGUGUGGGCGGGGUAAAUGCGUUUUUGAUGCUG---UUU- .....-...((((...((((((((((((........)))....-.(((((.(((...((.......))...))))))))))))))...))).....))))...---...- ( -27.50) >DroEre_CAF1 131067 96 + 1 UCCUUUUUAUAUUUUUCGCUACCCCGUUGAACUAAAAACUUUU-CGCCCAACGCCAU---------CGUUUGUGUGGGCGGGGUAAAUGCGUUUUUGAUGCUG---UUU- .........((((...((((((((((((........)))....-.(((((.(((...---------.....))))))))))))))...))).....))))...---...- ( -26.00) >DroYak_CAF1 135135 99 + 1 UCCUUUUUAUAUUUUUCGCUACCCCGUUGAACUAAAAACUUUU-UGCCCAACGCCAU---------CGUUUGUGUGGGCGGGGUAAAUGCGUUUUUGAUGCUGGUUUUU- .((....(((......((((((((((((........)))....-.(((((.(((...---------.....))))))))))))))...)))......)))..)).....- ( -26.10) >DroAna_CAF1 138052 83 + 1 UUAUU-U--UAUUUUU---------GUUGAACUAAAAACUUUUUUGGCCCAUGGUAUAG-----------AGGAGGGGGAGAGUAAAUGCGUUUUUGAUGCUG---UUU- ....(-(--(((((((---------......((((((....))))))(((.........-----------....))).))))))))).(((((...)))))..---...- ( -11.32) >consensus UCCUU_UUAUAUUUUUCGCUACCCCGUUGAACUAAAAACUUUU_CGCCCAACGCCAU_G_______CGUUUGUGUGGGCGGGGUAAAUGCGUUUUUGAUGCUG___UUU_ ...................(((((((((........)))......(((((.(((.................))))))))))))))...(((((...)))))......... (-18.08 = -19.20 + 1.11)
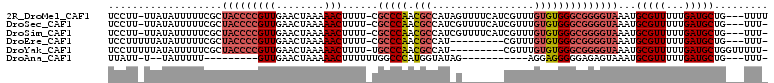
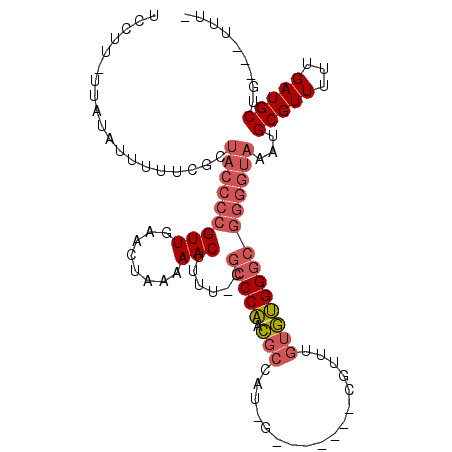
| Location | 13,829,866 – 13,829,971 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -13.94 |
| Energy contribution | -15.49 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13829866 105 - 20766785 AAAA---CAGCAUCAAAAACGCAUUUACCCCGCCCACACAAACGAUGAAAACUAUGGCGUUGGGCG-AAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAA-AAGGA ....---............(((...(((((((((.....................)))(((((((.-...........))))))))))))))))..........-..... ( -23.90) >DroSec_CAF1 130420 104 - 1 -AAA---CAGCAUCAAAAACGCAUUUACCCCGCCCACACAAACGAUGAAAACGAUGGCGUUGGGCG-AAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAA-AAGGA -...---............(((...((((((((((((.(...((.(....)))...).).))))).-....(((........))))))))))))..........-..... ( -24.10) >DroSim_CAF1 135671 104 - 1 -AAA---CAGCAUCAAAAACGCAUUUACCCCGCCCACACAAACGAUGAAAACGAUGGCGUUGGGCG-AAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAA-AAGGA -...---............(((...((((((((((((.(...((.(....)))...).).))))).-....(((........))))))))))))..........-..... ( -24.10) >DroEre_CAF1 131067 96 - 1 -AAA---CAGCAUCAAAAACGCAUUUACCCCGCCCACACAAACG---------AUGGCGUUGGGCG-AAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAAAAAGGA -...---............(((...(((((((((..........---------..)))(((((((.-...........))))))))))))))))................ ( -24.50) >DroYak_CAF1 135135 99 - 1 -AAAAACCAGCAUCAAAAACGCAUUUACCCCGCCCACACAAACG---------AUGGCGUUGGGCA-AAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAAAAAGGA -..................(((...(((((((((..........---------..)))(((((((.-(((....))).))))))))))))))))................ ( -24.40) >DroAna_CAF1 138052 83 - 1 -AAA---CAGCAUCAAAAACGCAUUUACUCUCCCCCUCCU-----------CUAUACCAUGGGCCAAAAAAGUUUUUAGUUCAAC---------AAAAAUA--A-AAUAA -...---..((.........))..................-----------........(((((.((((....)))).)))))..---------.......--.-..... ( -3.90) >consensus _AAA___CAGCAUCAAAAACGCAUUUACCCCGCCCACACAAACG_______C_AUGGCGUUGGGCG_AAAAGUUUUUAGUUCAACGGGGUAGCGAAAAAUAUAA_AAGGA ...................(((...(((((((((((........................)))))......(((........))))))))))))................ (-13.94 = -15.49 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:18 2006