
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,815,337 – 13,815,444 |
| Length | 107 |
| Max. P | 0.539006 |

| Location | 13,815,337 – 13,815,444 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -21.29 |
| Consensus MFE | -10.35 |
| Energy contribution | -10.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539006 |
| Prediction | RNA |
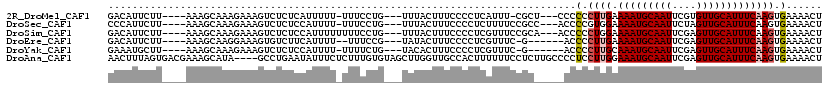
Download alignment: ClustalW | MAF
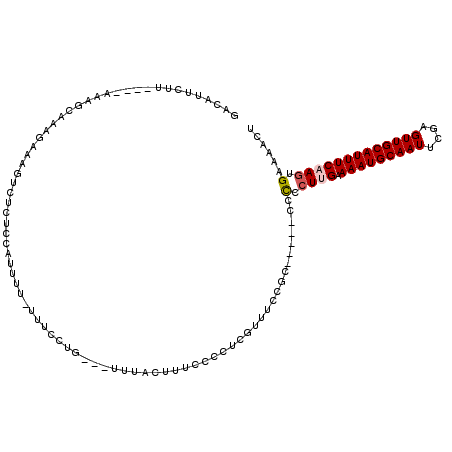
>2R_DroMel_CAF1 13815337 107 + 20766785 GACAUUCUU----AAAGCAAAGAAAGUCUCUCAUUUUU-UUUCCUG---UUUACUUUCCCCUCAUUU-CGCU---CCCCCCUUGAAAAUGCAAUUCGUGUUGCAUUUCAAGUGAAAACU (((.(((((----......))))).)))..........-.......---...............(((-((((---.......(((((.((((((....))))))))))))))))))... ( -20.01) >DroSec_CAF1 115860 108 + 1 CCCAUUCUU----AAAGCAAAGAAAGUCUCUCCAUUUU-UUUCCUG---UUUACUUUCCCCUCUUUUCCGCC---ACCCCGUGGAAAAUGCAAUUCUAGUUGCAUUUCAAGUGAAAACU ....(((((----......)))))..............-.......---(((((((..........(((((.---.....)))))(((((((((....))))))))).))))))).... ( -19.50) >DroSim_CAF1 121297 109 + 1 GACAUUCUU----AAAGCAAAGAAAGUCUCUCCAUUUUUUUUCCUG---UUUACUUUCCCCUCGUUUCCGCA---ACCCCCUGGAAAAUGCAAUUCGAGUUGCAUUUCAAGUGAAAACU (((.(((((----......))))).)))..................---(((((((..........((((..---......))))(((((((((....))))))))).))))))).... ( -20.20) >DroEre_CAF1 116209 103 + 1 GACAUUCUU----AAAGCAAGGAAAGUGUCUUCAUUUU--UUUCCG---UAUACUUUCCCCUCGUUUC-G------ACCCCUUGAAAAUGCAAUUCGAGUUGCAUUUCAAGUGAAAACU ....((((.----.((((..((((((((((........--.....)---.)))))))))....)))).-.------)...(((((((.((((((....))))))))))))).))).... ( -21.92) >DroYak_CAF1 120468 104 + 1 GAAAUGCUU----AAAGCAAAGAAAGUCUCUCCAUUUU-UUUUCUG---UACACUUUCCCCUCGUUUC-G------ACCCCUUGCAAAUGCAAUUCGAGUUGCAUUUCAAGUGAAAACU (((((((..----..((.(((((((((......)))))-)))))).---................(((-(------(....((((....)))).)))))..))))))).(((....))) ( -19.90) >DroAna_CAF1 122384 115 + 1 AACUUUAGUGACGAAAGCAUA----GCCUGAAUAUUUCUCUUUGUGUAGCUUGGUUGCCACUUUUUUCCUCUUGCCCCUCCUUGGAAAUGCAAUUCGAGUUGCAUUUCAAGUGAAAACU .......(((.(....)))).----.........((((.(((.(((((((((((((((......(((((..............))))).)))).)))))))))))...))).))))... ( -26.24) >consensus GACAUUCUU____AAAGCAAAGAAAGUCUCUCCAUUUU_UUUCCUG___UUUACUUUCCCCUCGUUUCCGC_____CCCCCUUGAAAAUGCAAUUCGAGUUGCAUUUCAAGUGAAAACU ..............................................................................(.((((.(((((((((....))))))))))))).)...... (-10.35 = -10.72 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:14 2006