
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,459,549 – 2,459,663 |
| Length | 114 |
| Max. P | 0.593235 |

| Location | 2,459,549 – 2,459,663 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -39.32 |
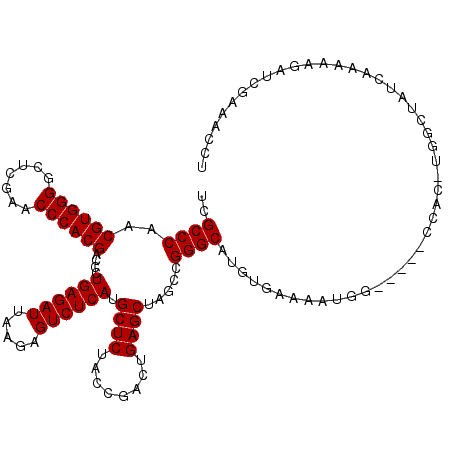
| Consensus MFE | -31.45 |
| Energy contribution | -31.45 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593235 |
| Prediction | RNA |
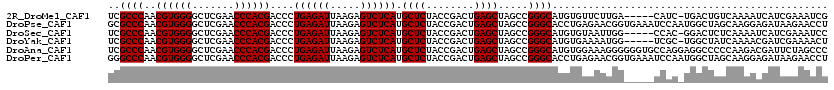
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2459549 114 + 20766785 UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGUUCUUGA-----CAUC-UGACUGUCAAAAUCAUCGAAAUCG ((((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))..(((...((((-----((..-....))))))...))).))..... ( -37.50) >DroPse_CAF1 31472 120 + 1 GCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACCUGAGAACGGUGAAAUCCAAUGGCUAGCAAGGAGAUAAGAACCU (((.....((((((.......))))))....((((((.....)))))))))(((..(..((..(((((((.((((((.......))))....))...))))))).))..)...))).... ( -39.40) >DroSec_CAF1 21640 114 + 1 UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGUAAUUGG-----CCAC-GGACUCUCAAAAUCAUCGAAAUCC (((.....((((((.......))))))....(((..((.(((((((...((((........)))).....((.((........)).-----))..-))))))).))..))).)))..... ( -33.90) >DroYak_CAF1 20455 114 + 1 UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGAAAAUGG-----UCGC-UGGCUAUCAAAACGAUCGAAAACU ((((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).(((....((((-----(...-..)))))....)))...))..... ( -36.00) >DroAna_CAF1 18075 120 + 1 UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGGAAAGGGGGGUGCCAGGAGGCCCCCAAGACGAUUCUAGCCC ..((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))...(((((.(.((((.(((....)))))))....)..))))).... ( -47.50) >DroPer_CAF1 22510 120 + 1 GGGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACCUGAGAACGGUGAAAUCCAAUGGCUAGCAAGGAGAUAAGAACCU ((((....((((((.......))))))....((((((.....)))))).))))...(..((..(((((((.((((((.......))))....))...))))))).))..).......... ( -41.60) >consensus UCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUGUGAAAAUGG_____CCAC_UGGCUAUCAAAAAGAUCGAAACCU ..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).............................................. (-31.45 = -31.45 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:09 2006