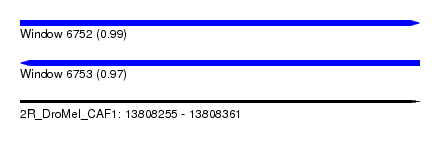
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,808,255 – 13,808,361 |
| Length | 106 |
| Max. P | 0.993708 |

| Location | 13,808,255 – 13,808,361 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 71.24 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -19.13 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
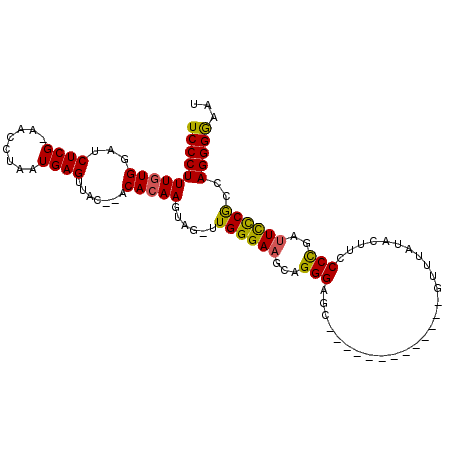
>2R_DroMel_CAF1 13808255 106 + 20766785 UCCCUUUCUGGAUCUCGCCAUCUAAUGAGUUGCACACACAAGUUG-UUGGGAUGCAGGGAGCCAUCGAUUCGGUUGUUUAUACUUCCCCGAUUUUCGUCAGGGGAAU ((((((..(((.((((((..(((((..(.(((......))).)..-)))))..)).)))).))).(((.((((..((....))....))))...)))..)))))).. ( -30.20) >DroEre_CAF1 109291 92 + 1 UCCCUUUGUGGAUCUCG-AACUUAAUGAGUUAC--ACACAAGUAGAUUGGGAAGUAGGGAGCU------------GUUUAUACUUCCCUGAUUCCCACCAGGGAAUU (((((.((((...((((-.......))))...)--))).........((((((.(((((((..------------((....))))))))).))))))..)))))... ( -30.20) >DroYak_CAF1 113535 82 + 1 UCCCUUUGUGGAUCUCG-AACCUAAUGAGUUAC--ACACAAGUAGUUUGGGAAGCAGGGAGA----------------------GCCCCGAUUCCCGCCAGGGGAAU ((((((((((...((((-.......))))...)--)))..........(((((...(((...----------------------..)))..)))))...)))))).. ( -26.80) >consensus UCCCUUUGUGGAUCUCG_AACCUAAUGAGUUAC__ACACAAGUAG_UUGGGAAGCAGGGAGC_____________GUUUAUACUUCCCCGAUUCCCGCCAGGGGAAU ((((((((((...((((........)))).......)))))......((((((...(((...........................)))..))))))..)))))... (-19.13 = -18.70 + -0.43)

| Location | 13,808,255 – 13,808,361 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 71.24 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -16.91 |
| Energy contribution | -18.36 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
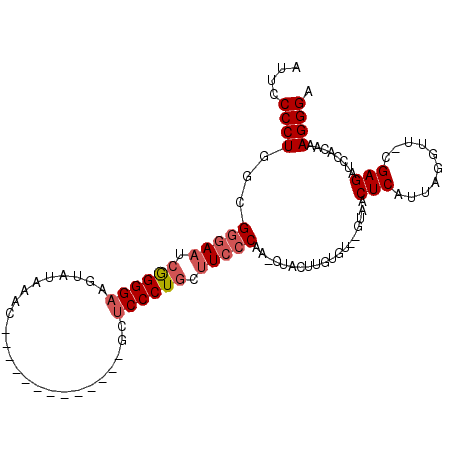
>2R_DroMel_CAF1 13808255 106 - 20766785 AUUCCCCUGACGAAAAUCGGGGAAGUAUAAACAACCGAAUCGAUGGCUCCCUGCAUCCCAA-CAACUUGUGUGUGCAACUCAUUAGAUGGCGAGAUCCAGAAAGGGA ....((((..(((...((((..............)))).))).((((((((((((..((((-....))).)..))))..((....)).)).)))..)))...)))). ( -23.14) >DroEre_CAF1 109291 92 - 1 AAUUCCCUGGUGGGAAUCAGGGAAGUAUAAAC------------AGCUCCCUACUUCCCAAUCUACUUGUGU--GUAACUCAUUAAGUU-CGAGAUCCACAAAGGGA ...(((((.(((((..((.((((((((.....------------.......)))))))).....(((((.((--(.....)))))))).-.))..)))))..))))) ( -28.80) >DroYak_CAF1 113535 82 - 1 AUUCCCCUGGCGGGAAUCGGGGC----------------------UCUCCCUGCUUCCCAAACUACUUGUGU--GUAACUCAUUAGGUU-CGAGAUCCACAAAGGGA ....((((...(((((.(((((.----------------------...))))).))))).......(((((.--(...(((........-.)))..)))))))))). ( -27.00) >consensus AUUCCCCUGGCGGGAAUCGGGGAAGUAUAAAC_____________GCUCCCUGCUUCCCAA_CUACUUGUGU__GUAACUCAUUAGGUU_CGAGAUCCACAAAGGGA ....((((...(((((.((((((........................)))))).)))))...................(((..........)))........)))). (-16.91 = -18.36 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:10 2006