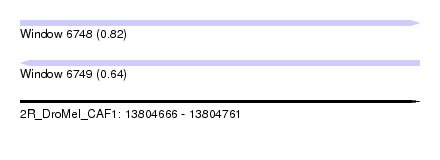
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,804,666 – 13,804,761 |
| Length | 95 |
| Max. P | 0.823723 |

| Location | 13,804,666 – 13,804,761 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -13.88 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
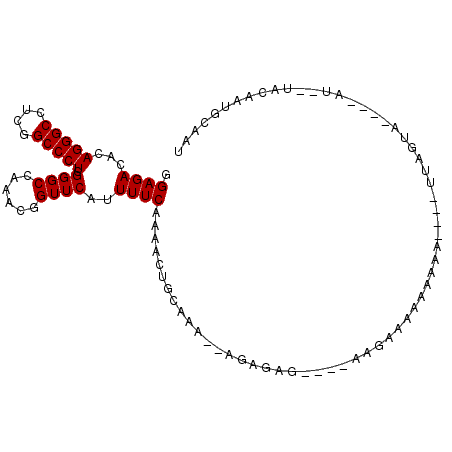
>2R_DroMel_CAF1 13804666 95 + 20766785 GGAGACACAGGGCCUCGGCCCUCGGGCCAAAUGGUUCAUUUUCAAAACUGCAAG--AUAGAG----CAGAGCGAACGAUUGAUUAGUU----GU--UGCAAUGCAAU (((((...(((((....))))).(((((....))))).)))))....((((...--.....)----))).((.((((((......)))----))--)))........ ( -28.50) >DroPse_CAF1 102469 98 + 1 GGAGACGCAGGGCCUCGGCCCUCGGGCCAAACUGUUCAUUUUCAAAACUGCAAA---GAGAG--CGGAGAAACAAAAA----UUAAUAUGGAAUAUAAGAAUAUAAU ((...((.(((((....)))))))..))...((((((.((((((....)).)))---).)))--)))...........----......................... ( -22.80) >DroWil_CAF1 128676 90 + 1 AGAGACACAGGGCCUCGGCCCUCGGGCCAAACUGUUCAUUUUCAAAACUGCAAA--AGCAAA----AAGAAAACAAAA----AUAGUC----AA---GAAAUUAAAU ...(((..(((((....))))).((((......)))).(((((.....(((...--.)))..----..))))).....----...)))----..---.......... ( -17.80) >DroYak_CAF1 109948 92 + 1 GGAGACACAGGGCCUCGGCCCUCGGGCCAAAUGGUUCAUUUUCAAAACUGCAAG--AUAGAGAACGUAGUAGG---GA----UUAGUU----GU--UGCAAUGCAAU (((((...(((((....))))).(((((....))))).))))).....((((..--...((.(((.((((...---.)----))))))----.)--)....)))).. ( -23.60) >DroAna_CAF1 113882 91 + 1 UGAGACACAGGGCCUCGGCCCUCGGGCCAAAUGGUUCAUUUUCAAAACUGCAAUUAUUCGAG----AGGAGAAUAAGA----UUCG------AU--UAAUAUGCAAU (((((...(((((....))))).(((((....)))))..)))))....((((((((.(((((----............----))))------).--)))).)))).. ( -25.70) >DroPer_CAF1 103447 96 + 1 GGAGACGCAGGGCCUCGGCCCUCGGGCCAAACUGUUCAUUUUCAAAACUGCAAA---GAGAG--CGAAGAAACAAAAA----UUAAUAUGGAAU--AACAAUAUAAU ((...((.(((((....)))))))..))....(((((.((((((....)).)))---).)))--))............----............--........... ( -19.50) >consensus GGAGACACAGGGCCUCGGCCCUCGGGCCAAACGGUUCAUUUUCAAAACUGCAAA__AGAGAG____AAGAAAAAAAAA____UUAGUA____AU__UACAAUGCAAU .((((...(((((....))))).((((......))))..))))................................................................ (-13.88 = -13.88 + 0.00)

| Location | 13,804,666 – 13,804,761 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -14.80 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
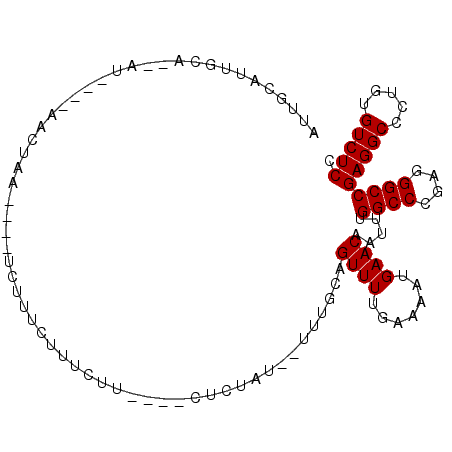
>2R_DroMel_CAF1 13804666 95 - 20766785 AUUGCAUUGCA--AC----AACUAAUCAAUCGUUCGCUCUG----CUCUAU--CUUGCAGUUUUGAAAAUGAACCAUUUGGCCCGAGGGCCGAGGCCCUGUGUCUCC ...(((..((.--..----..........(((((..(.(((----(.....--...))))....)..)))))....(((((((....)))))))))..)))...... ( -23.40) >DroPse_CAF1 102469 98 - 1 AUUAUAUUCUUAUAUUCCAUAUUAA----UUUUUGUUUCUCCG--CUCUC---UUUGCAGUUUUGAAAAUGAACAGUUUGGCCCGAGGGCCGAGGCCCUGCGUCUCC .........................----.............(--(....---...)).((((.......)))).....((((....))))(((((.....))))). ( -16.60) >DroWil_CAF1 128676 90 - 1 AUUUAAUUUC---UU----GACUAU----UUUUGUUUUCUU----UUUGCU--UUUGCAGUUUUGAAAAUGAACAGUUUGGCCCGAGGGCCGAGGCCCUGUGUCUCU ..........---..----(((...----..(..(((((..----.((((.--...))))....)))))..)(((((((((((....)))))))...)))))))... ( -24.80) >DroYak_CAF1 109948 92 - 1 AUUGCAUUGCA--AC----AACUAA----UC---CCUACUACGUUCUCUAU--CUUGCAGUUUUGAAAAUGAACCAUUUGGCCCGAGGGCCGAGGCCCUGUGUCUCC ...(((..((.--..----......----..---........((((....(--(..........))....))))..(((((((....)))))))))..)))...... ( -17.10) >DroAna_CAF1 113882 91 - 1 AUUGCAUAUUA--AU------CGAA----UCUUAUUCUCCU----CUCGAAUAAUUGCAGUUUUGAAAAUGAACCAUUUGGCCCGAGGGCCGAGGCCCUGUGUCUCA ((((((.((((--.(------(((.----............----.)))).))))))))))..................((((....))))(((((.....))))). ( -23.44) >DroPer_CAF1 103447 96 - 1 AUUAUAUUGUU--AUUCCAUAUUAA----UUUUUGUUUCUUCG--CUCUC---UUUGCAGUUUUGAAAAUGAACAGUUUGGCCCGAGGGCCGAGGCCCUGCGUCUCC .....((((((--...........(----((((((...((..(--(....---...))))...))))))).))))))..((((....))))(((((.....))))). ( -17.80) >consensus AUUGCAUUGCA__AU____AACUAA____UCUUUCUUUCUU____CUCUAU__UUUGCAGUUUUGAAAAUGAACAAUUUGGCCCGAGGGCCGAGGCCCUGUGUCUCC ...........................................................((((.......)))).....((((....))))(((((.....))))). (-14.80 = -14.80 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:06 2006