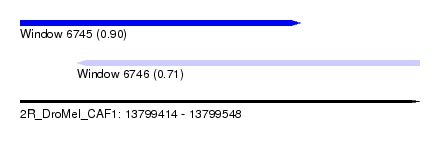
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,799,414 – 13,799,548 |
| Length | 134 |
| Max. P | 0.900271 |

| Location | 13,799,414 – 13,799,508 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 66.33 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13799414 94 + 20766785 AUUUAUUUCUGCUUUCAAAAGCCUA---AAACUGAAUUCUGUAUGCUUUACUUACCGAACUUCUGGUCUCGGUGAAGUUGCUCAGGUCAGGAGUGAG ..((((((..((((....))))...---...((((..((((...((...(((((((((((.....)).)))))).))).)).)))))))))))))). ( -22.40) >DroSec_CAF1 100189 94 + 1 AUUCAUUUCGGCUUUUAAAAGAAAA---CAACUCAGUCCUGUAUGUUUUACUUACCGAACUUCUGGUCUCGGUGAAGUUGCUCAGGUCAGGAGUGAG .....((((...........)))).---...((((.(((((..((....(((((((((((.....)).)))))).)))....))...))))).)))) ( -25.70) >DroSim_CAF1 104062 94 + 1 AUUCAUUUCGUCUUUCAAAAGAAGA---CAACUCAAUCCUGUAUGUUUUACUUACCGAACUUCUGGUCUCGGUGAAGUUGCUCAGGUCAGGAGUGAG .........((((((.....)))))---)..((((.(((((..((....(((((((((((.....)).)))))).)))....))...))))).)))) ( -28.70) >DroEre_CAF1 100622 72 + 1 AU----------------------A---CAAUUCAAUUCUGUCUGCUUUACUUACCGAACUUCUGGUCUCGGUAAAGUUGCUCAGGUCGGGAGUGAG ..----------------------.---....(((.(((((((((....(((((((((((.....)).))))).))))....)))).))))).))). ( -20.20) >DroWil_CAF1 123564 82 + 1 ----AG--------UUAAAGGGAAUCGCCAACCGAG---AGUAGAGAAUACAUACCGAACUUCUGGUUUCGGUGAAAUUGCUCAGAUCGGGCGUUAA ----..--------...........((((.((((((---(.(((((..............))))).)))))))((......))......)))).... ( -18.94) >DroAna_CAF1 108994 71 + 1 ------------------AGGACUA---GGAAG-GAUUAGAGAUG----UCUUACCGAACUUCUGGUCUCGGUAAAGUUGCUGAGGUCCGGUGUCAG ------------------..(((..---((.((-(((.......)----)))).))......((((((((((((....)))))))).)))).))).. ( -22.30) >consensus AU__AU________U_AAAAGAAAA___CAACUCAAUUCUGUAUGCUUUACUUACCGAACUUCUGGUCUCGGUGAAGUUGCUCAGGUCAGGAGUGAG ..........................................................(((((((..((.(((......))).))..)))))))... (-12.40 = -12.60 + 0.20)

| Location | 13,799,433 – 13,799,548 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.63 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13799433 115 - 20766785 AACUCCAAAGGGACGCGGAAAGCGCGGACGCAGUGCAGGUCUCACUCCUGACCUGAGCAACUUCACCGAGACCAGAAGUUCGGUAAGUAAAGCAUACAGAAUUCAGUUUUAGGCU ....((....(..((((.....))))..)((.((.((((((........)))))).)).((((.((((((........))))))))))...))..................)).. ( -32.70) >DroVir_CAF1 122566 93 - 1 AACUCCAAAGGGACGCGGCAAGCGCGGACGCAGCGCUGGCUUAACGCCCGAUCUGAGCAACUUUACCGAAACCAGAAGCUCGGUGAGUCGA----------------------CU .((((....(((.((.(((.(((((.......))))).)))...)))))...((((((..((...........))..)))))).))))...----------------------.. ( -29.10) >DroEre_CAF1 100624 110 - 1 AACUCCAAAGGGACGCGGCAAGCGCGGACGCAGUGCAGGUCUCACUCCCGACCUGAGCAACUUUACCGAGACCAGAAGUUCGGUAAGUAAAGCAGACAGAAUUGAAUUGU----- .....(((..(..((((.....))))..)((.((.((((((........)))))).)).((((.((((((........))))))))))...))........)))......----- ( -31.40) >DroYak_CAF1 104796 108 - 1 AACUCCAAAGGGACGCGGCAAGCGCGGACGCAGUGCAGGUCUCACUCCCGACCUGAGCAACUUCACCGAAACCAGAAGUUCGGUAAGUAAAACAAACAGGAUUUAA--GA----- ...(((....(..((((.....))))..)...((.((((((........)))))).)).((((.((((((........))))))))))..........))).....--..----- ( -31.40) >DroMoj_CAF1 136453 90 - 1 UACUCCAAAGGGACGCGGCAAGCGCGGACGCAGCGCAGGUCUGACGCCCGAUCUGAGCAACUUUACCGAAACCAGAAGCUCGGUGAGUAC------------------------- (((((....(((.((((((..((((.......))))..).))).)))))...((((((..((...........))..)))))).))))).------------------------- ( -29.80) >DroAna_CAF1 108995 110 - 1 AACUCCAAAGGGACGCGGCAAGCGCGGACGCAGUGCAGGUCUGACACCGGACCUCAGCAACUUUACCGAGACCAGAAGUUCGGUAAGA----CAUCUCUAAUC-CUUCCUAGUCC ..........(((((((.....)))(((.((..((.(((((((....)))))))))))...(((((((((........))))))))).----.........))-)......)))) ( -32.70) >consensus AACUCCAAAGGGACGCGGCAAGCGCGGACGCAGUGCAGGUCUCACUCCCGACCUGAGCAACUUUACCGAAACCAGAAGUUCGGUAAGUAAA_CA_ACAGAAUU_A____U_____ ..........(..((((.....))))..)...((.((((((........)))))).)).((((.((((((........))))))))))........................... (-25.34 = -25.45 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:03 2006