| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,797,656 – 13,797,767 |
| Length | 111 |
| Max. P | 0.695465 |

| Location | 13,797,656 – 13,797,767 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -50.03 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
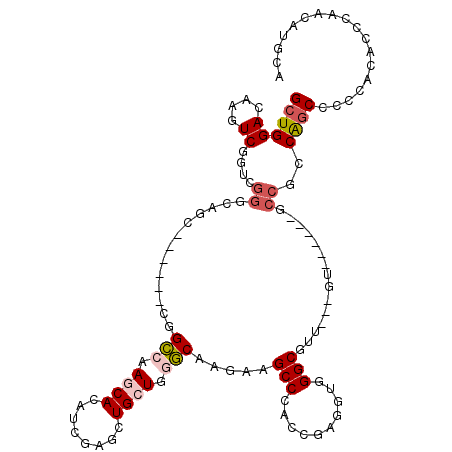
>2R_DroMel_CAF1 13797656 111 - 20766785 GCUGGACAAGUCAGUCGGCCAGGCGGUGACGGCAAAGCACAUCGAAUUGAUGGGCAAGAAGCCGACGGAAGUGGGCGUUGGUGU------UCCGCCACCACCAGCUCCCAAUAUGUA (((((....(((....)))..(((((.((((.(((.((.((((.....)))).)).....(((.((....)).))).))).)))------))))))....)))))............ ( -44.20) >DroVir_CAF1 120820 93 - 1 GCUGGACAAGUCG------CAGC------CGGGCAAGCACAUUGAGCUGCUGGCCAAGAAGCCCAGCGAGGUGGGCG------U------GCCGGCAGCGCCCACGCCCAAUAUACC ((((((....)).------))))------.((((.(((.......)))(((((.(.....).)))))...(((((((------(------.......))))))))))))........ ( -40.70) >DroPse_CAF1 95786 102 - 1 GCUGGACAAGUCGGUUGGGCAAC------CGGCCAAGCACAUCGAGCUGCUCGGCAAGAAGCCCCCCGAGGUGGGCGUG---GU------GCCGGCAGCUCCCACACCCAGCAUGCC (((((.......((((((....)------))))).........(((((((.(((((....((((((...)).))))...---.)------)))))))))))......)))))..... ( -55.30) >DroSim_CAF1 102328 111 - 1 GCUGGACAAGUCAGUCGGCCAGGCGGUGACGGCGAAGCACAUCGAAUUGAUGGGCAAGAAGCCGACCGAAGUGGGCGUUGGUGU------UCCGCCAGCCCCCGCACCCAACAUGUA (.(((.....((.((((((..(.((....)).)...((.((((.....)))).)).....)))))).)).(((((.(((((((.------..))))))).)))))..))).)..... ( -46.20) >DroAna_CAF1 107244 117 - 1 GCUGGACAAGUCGGUCGGGCAGGCGGCGGCGGCCAAGCACAUCGAGCUGCUGGGCAAGAAGCCGCCGGAGGUGGGCGUUGGUGUUGGAGUGCCUCCGGCCCCAGCUCCCAACAUGUA (((.(((......))).)))...(((((((.(((.((((........)))).))).....)))))))..(.((((.(((((.(((((((...))))))).))))).)))).)..... ( -58.50) >DroPer_CAF1 96653 102 - 1 GCUGGACAAGUCGGUUGGGCAAC------CGGCCAAGCACAUCGAGCUGCUCGGCAAGAAGCCCCCCGAGGUGGGCGUG---GU------GCCGGCAGCUCCCACACCCAGCAUGCC (((((.......((((((....)------))))).........(((((((.(((((....((((((...)).))))...---.)------)))))))))))......)))))..... ( -55.30) >consensus GCUGGACAAGUCGGUCGGGCAGC______CGGCCAAGCACAUCGAGCUGCUGGGCAAGAAGCCCACCGAGGUGGGCGUU___GU______GCCGCCAGCCCCCACACCCAACAUGCA ((((((....))....((.............(((.((((........)))).))).....(((..........)))...............))..)))).................. (-15.10 = -15.85 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:58 2006