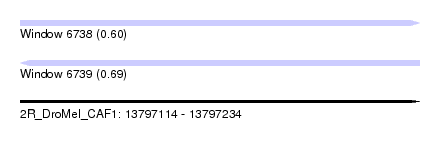
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,797,114 – 13,797,234 |
| Length | 120 |
| Max. P | 0.688215 |

| Location | 13,797,114 – 13,797,234 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -57.83 |
| Consensus MFE | -32.29 |
| Energy contribution | -30.60 |
| Covariance contribution | -1.69 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
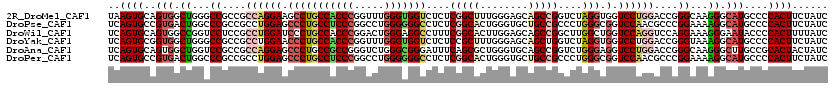
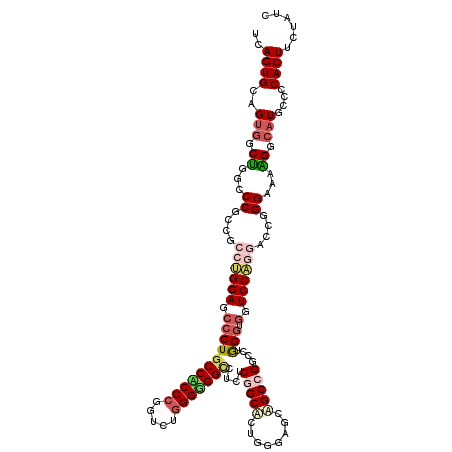
>2R_DroMel_CAF1 13797114 120 + 20766785 GAUAGAAGUGGGGCAUGCCCUUGCCCGGUCCAGGACCACCUAGACCGGCUGCUCCCAAAGCCGAGAGACCACCCAAACCGGGUGGCAGGCUUCCUGGCGGCGGCCCAGCCACUGCACUUA .....((((((((((......)))))((((.(((....))).))))(((((((.((((((((....(.((((((.....))))))).)))))..))).))))))).........))))). ( -58.10) >DroPse_CAF1 95205 120 + 1 GAUAGAAGUGGGGCAUGCCUUUUCCGGGCGUUGGACCGCCCAGGGCGGCAGCACCCAGUGCCGAGAGGCCCCCCAGGCCGGGAGGCAGGGCUCCAGGCGGCGGGCCAGUCACGGCACUGA .........(((((.((((...((((((((......))))).))).)))))).)))(((((((.(((((((.((..(((.((((......)))).))))).)))))..)).))))))).. ( -64.40) >DroWil_CAF1 121227 120 + 1 GAUAAAAGUGGGGUAUUCCCUUUCCUGGACCUGGACCAGCCAAGCCGGCUGCUCCAAGUGCCGAAAGGCCUCCCAGUCCGGGUGGCAGGGAUCCAGGCGGAGGACCGGCCACUGCACUGA ......((((.(((..(((((..(((((((.((((.(((((.....))))).))))((.(((....)))))....)))))))....)))))....(((((....)).)))))).)))).. ( -55.30) >DroYak_CAF1 102522 120 + 1 GAUAGAAGUGGGGCAUGCCUUUACCCGGUCCAGGACCACCUAGACCAGCUGCUCCCAAAGCGGAGAGACCACCCAAACCGGGUGGCAGGGUUCCAGGCGGCGGCCCAGCCACCGCACUGA ......((((.((...((((..((((((((.(((....))).))))..(((((.....)))))...(.((((((.....))))))).))))...))))(((......))).)).)))).. ( -53.10) >DroAna_CAF1 106690 120 + 1 GAUAGUAGUGCGGCAAGCCCUUGCCCGGUCCAGGACCUCCCAGACCGGCUGCACCCAGCGCUGAAAUCCCGCCCAGACCCGGCGGCAGGGCUCCUGGCGGCGGACCAGCCACUGCACUGA ....((((((((((((....))))).(((((.((.....))......(((((....(((.(((.....(((((.......)))))))).)))....)))))))))).).))))))..... ( -51.70) >DroPer_CAF1 96066 120 + 1 GAUAGAAGUGGGGCAUGCCUUUUCCGGGCGUUGGACCGCCCAGGGCGGCAGCACCCAGUGCCGAGAGGCCCCCCAGGCCGGGAGGCAGGGCUCCAGGCGGCGGGCCAGUCACGGCACUGA .........(((((.((((...((((((((......))))).))).)))))).)))(((((((.(((((((.((..(((.((((......)))).))))).)))))..)).))))))).. ( -64.40) >consensus GAUAGAAGUGGGGCAUGCCCUUUCCCGGUCCAGGACCACCCAGACCGGCUGCACCCAGUGCCGAGAGGCCACCCAGACCGGGUGGCAGGGCUCCAGGCGGCGGACCAGCCACUGCACUGA ......((((.((...(((((((((.(((...((.....))....(((((........))))).............))).))))..))))).)).(((((....)).)))....)))).. (-32.29 = -30.60 + -1.69)

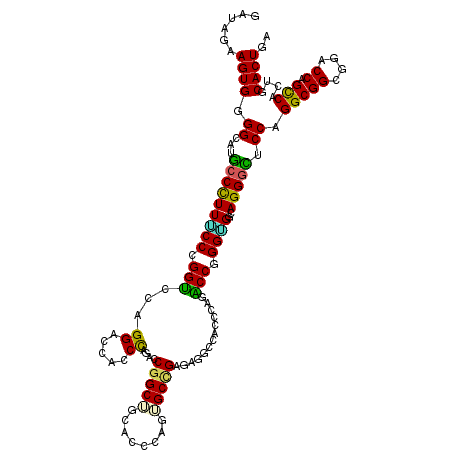
| Location | 13,797,114 – 13,797,234 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -61.82 |
| Consensus MFE | -32.39 |
| Energy contribution | -33.28 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13797114 120 - 20766785 UAAGUGCAGUGGCUGGGCCGCCGCCAGGAAGCCUGCCACCCGGUUUGGGUGGUCUCUCGGCUUUGGGAGCAGCCGGUCUAGGUGGUCCUGGACCGGGCAAGGGCAUGCCCCACUUCUAUC ...(.((.(((((...))))).)))..((((((.(((((((.....))))))).....))))))(((.(((.((((((((((....))))))))))((....)).))))))......... ( -58.10) >DroPse_CAF1 95205 120 - 1 UCAGUGCCGUGACUGGCCCGCCGCCUGGAGCCCUGCCUCCCGGCCUGGGGGGCCUCUCGGCACUGGGUGCUGCCGCCCUGGGCGGUCCAACGCCCGGAAAAGGCAUGCCCCACUUCUAUC ..(((((((.((..(((((.(((((.((((......)))).)))..)).))))))).)))))))(((.((((((...(((((((......)))))))....)))).)))))......... ( -69.90) >DroWil_CAF1 121227 120 - 1 UCAGUGCAGUGGCCGGUCCUCCGCCUGGAUCCCUGCCACCCGGACUGGGAGGCCUUUCGGCACUUGGAGCAGCCGGCUUGGCUGGUCCAGGUCCAGGAAAGGGAAUACCCCACUUUUAUC .......(((((..(((..(((.((((((((((..((....))...)))).(((....))).((((((.(((((.....))))).))))))))))))....)))..))))))))...... ( -53.50) >DroYak_CAF1 102522 120 - 1 UCAGUGCGGUGGCUGGGCCGCCGCCUGGAACCCUGCCACCCGGUUUGGGUGGUCUCUCCGCUUUGGGAGCAGCUGGUCUAGGUGGUCCUGGACCGGGUAAAGGCAUGCCCCACUUCUAUC ((((.((((((((...))))))))))))......(((((((.....)))))))..........((((.(((.((((((((((....))))))))))((....)).)))))))........ ( -57.90) >DroAna_CAF1 106690 120 - 1 UCAGUGCAGUGGCUGGUCCGCCGCCAGGAGCCCUGCCGCCGGGUCUGGGCGGGAUUUCAGCGCUGGGUGCAGCCGGUCUGGGAGGUCCUGGACCGGGCAAGGGCUUGCCGCACUACUAUC .((((((..((((.((....))))))(((..((((((..........))))))..))).))))))(((((...(((((..((....))..)))))(((((....))))))))))...... ( -61.60) >DroPer_CAF1 96066 120 - 1 UCAGUGCCGUGACUGGCCCGCCGCCUGGAGCCCUGCCUCCCGGCCUGGGGGGCCUCUCGGCACUGGGUGCUGCCGCCCUGGGCGGUCCAACGCCCGGAAAAGGCAUGCCCCACUUCUAUC ..(((((((.((..(((((.(((((.((((......)))).)))..)).))))))).)))))))(((.((((((...(((((((......)))))))....)))).)))))......... ( -69.90) >consensus UCAGUGCAGUGGCUGGCCCGCCGCCUGGAGCCCUGCCACCCGGUCUGGGGGGCCUCUCGGCACUGGGAGCAGCCGGCCUGGGUGGUCCAGGACCGGGAAAAGGCAUGCCCCACUUCUAUC ..((((..(((.((...((....((((((.(((((((((((.....)))))))....(((((........)))))....))).).))))))....))...)).)))....))))...... (-32.39 = -33.28 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:57 2006