| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,796,675 – 13,796,791 |
| Length | 116 |
| Max. P | 0.773042 |

| Location | 13,796,675 – 13,796,791 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
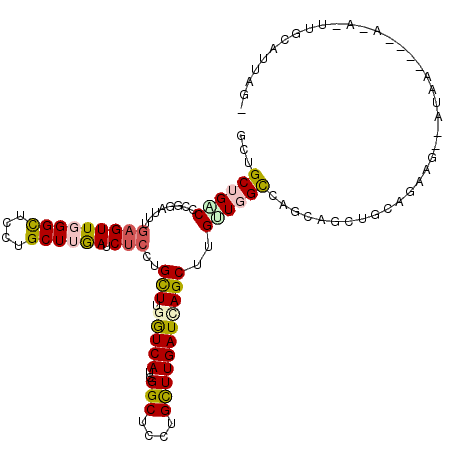
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.773042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
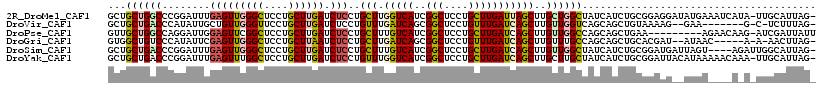
>2R_DroMel_CAF1 13796675 116 + 20766785 GCUGCUGGCCCGGAUUUGAGUUGGGCUCCUGCUUGAUCUCCUGCUUGGUCAUCGGCUCCUGCUUGAUUAGCUUGCUGGCUAUCAUCUGCGGAGGAUAUGAAAUCAUA-UUGCAUUAG- ((((.(((((.((....((((..(((....)))..).)))...)).))))).))))..((((.((((.((((....))))))))...))))..((((((....))))-)).......- ( -39.70) >DroVir_CAF1 119780 106 + 1 GCUGCUGACCCAUAUUGCUGUUGGGUUCCUGCUUGAUCUCCUGUUUGAUCAGCGGCUCCUGUUUGAUCAGCUUGUUGGUCAGCAGCUGUAAAAG--GAA-------G-C-UCUUUAG- ((((((((((.(((..((((.((((...(((((.((((........)))))))))..))))......)))).))).))))))))))....((((--(..-------.-.-)))))..- ( -41.10) >DroPse_CAF1 94785 108 + 1 GUUGCUGGCCAGGAUUGGAGUUCGGCUCCUGCUUGAUCUCCUGCUUUGUCAUCGGCUCCUGCUUGAUCAGCUUGUUGGCCAGCAGCUGAA---------AGAACAAG-AUCGAUUAUU ((((((((((((.(..((((.(((((....)).))).)))).(((..((((..(((....))))))).))).).))))))))))))....---------........-.......... ( -39.70) >DroGri_CAF1 115517 108 + 1 GUGGCUGUCCCAUAUUCGAGUUGGGCUCCUGCUUAAUCUCCUGCUUGAUCAGCGGCUCCUGUUUGAUCAGCUUGUUUGCCAGCAGCUGCACGAU--AUAAC-----A-A-AACUUAG- (..(((((.........(((((((((....)))))).)))..(((.(((((((((...))).)))))))))..........)))))..).....--.....-----.-.-.......- ( -29.90) >DroSim_CAF1 101350 113 + 1 GCUGCUGACCCGGAUUUGAGUUGGGCUCCUGCUUGAUCUCCUGCUUUGUCAUCGGCUCCUGCUUGAUCAGCUUGUUGGCUAUCAUCUGCGGAUGAUUAGU----AGAUUGGCAUUAG- ..(((..(.........((((..(((....)))..).)))(((((..((((((.((.......((((.((((....))))))))...)).)))))).)))----)).)..)))....- ( -36.10) >DroYak_CAF1 102087 116 + 1 GCUGCUGACCCGGAUUUGAGUUUGGCUCCUGCUUGAUCUCCUGUUUGGUCAUCGGCUCCUGCUUGAUCAGCUUGCUUGCUAUCAUCUGCGGAUUACAUAAAAACAAA-UUGCAUUAG- ((((.(((((.((((..(((((.(((....))).)).)))..))))))))).))))..((((.((((.(((......)))))))...))))................-.........- ( -32.50) >consensus GCUGCUGACCCGGAUUUGAGUUGGGCUCCUGCUUGAUCUCCUGCUUGGUCAUCGGCUCCUGCUUGAUCAGCUUGUUGGCCAGCAGCUGCAGAAG__AUAA____A_A_UUGCAUUAG_ ...((((((........(((((((((....)))))).)))..(((.(((((..(((....)))))))))))..))))))....................................... (-17.19 = -17.50 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:55 2006