| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
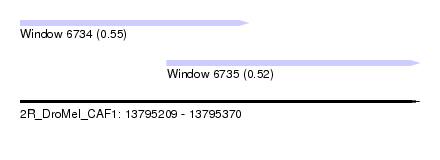
| Location | 13,795,209 – 13,795,370 |
| Length | 161 |
| Max. P | 0.547650 |

| Location | 13,795,209 – 13,795,301 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -19.36 |
| Energy contribution | -18.42 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13795209 92 + 20766785 CCUGCACCGCCGCCUCCCGCUGCGCCAUUGCCGGCAGCGCCUCCCACACCGCCAACGGCUCCAUUGCCCGAUGGCCCGUUCGGUGGCGGACC ......((((((((...((((((((....))..))))))...........((((.((((......).))).))))......))))))))... ( -38.70) >DroPse_CAF1 93279 92 + 1 CCUGCGCCACCUCCACCAGCAGGGCCAUUCACAGCGGCUCCACCGACUCCUCCCACACCGCCGUUGCCGGGUGGUCCACUGGGUGGUGGGCC ...((.(((((....((((..(((((((((.(((((((.....................)))))))..))))))))).))))..))))))). ( -41.60) >DroEre_CAF1 96438 92 + 1 CCUGCACCGCCGCCACCCGCUGCGCCAUUACCGGCAGCGCCUCCCACUCCGCCCACGGCUCCAUUGCCCGAUGGCCCGUUCGGUGGCGGACC ......((((((((...((((((..........))))))...............((((..(((((....))))).))))..))))))))... ( -36.80) >DroYak_CAF1 100682 92 + 1 CCUGCACCGCCGCCACCCGCUGCGCCAUUACCGGCAGCGCCUCCCACUCCGCCCACGGCUCCAUUGCCCGAUGGCCCGUUCGGUGGCGGACC ......((((((((...((((((..........))))))...............((((..(((((....))))).))))..))))))))... ( -36.80) >DroAna_CAF1 104826 92 + 1 CCUGCACCGCCUCCUCCCGGAGCUCCGUUGCCGGCCGCUCCGCCCACACCACCGACGGCUCCGUUACCAGAUGGCCCGUUUGGCGGUGGUCC ....(((((((......((((((.(((....)))..))))))...........(((((..(((((....))))).))))).))))))).... ( -37.20) >DroPer_CAF1 94145 92 + 1 CCUGCGCCACCUCCACCAGCAGGGCCAUUCACAGCGGCUCCACCGACUCCUCCCACACCGCCGUUGCCGGGUGGUCCACUGGGUGGUGGGCC ...((.(((((....((((..(((((((((.(((((((.....................)))))))..))))))))).))))..))))))). ( -41.60) >consensus CCUGCACCGCCGCCACCCGCAGCGCCAUUACCGGCAGCGCCUCCCACUCCGCCCACGGCUCCAUUGCCCGAUGGCCCGUUCGGUGGCGGACC ......(((((.((.......((((...........))))....................(((((....))))).......)).)))))... (-19.36 = -18.42 + -0.94)

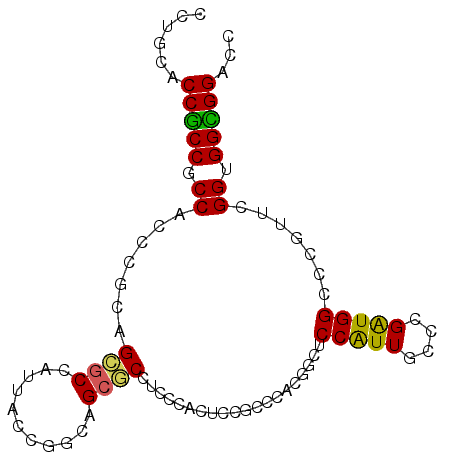
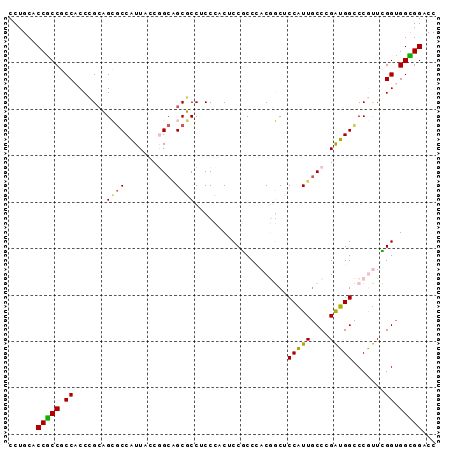
| Location | 13,795,268 – 13,795,370 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.34 |
| Mean single sequence MFE | -48.28 |
| Consensus MFE | -26.86 |
| Energy contribution | -25.83 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13795268 102 + 20766785 UCCAUUGCCCGAUGGCCCGU------UCGGUGGCGGACCUCCGGGCAUCGACUGCUUGUGGCCGUCCAAUGACGAGCCGGGCGUCACUCCGCCAUGUCCUUUCGGGCC ......((((((((((.(((------(...((((((.((..((((((.....)))))).))))).)))..)))).))))((((......))))........)))))). ( -45.50) >DroPse_CAF1 93338 99 + 1 GCCGUUGCCGGGUGGUCCAC------UGGGUGGUGGGCCACCAGGCAUCGUCUGUUUGUGCGCGUCUAGCGCUGAGCCAGGCGGCACUCCG---UGCGGCUUCUGGCC (((..((((.((((((((((------(....))))))))))).)))).((((((((((.((((.....)))))))).))))))((((...)---))))))........ ( -53.40) >DroSec_CAF1 96089 102 + 1 UCCAUUGCCCGAUGGCCCGU------UCGGUGGCGGACCUCCGGGCAUCGACUGCUUGUGGCCGUCUAAUGACGAGCCGGGCGUCACUCCGCCAUGUCCUUUCGGGCC ......((((((.((..(((------..(((((.(((((((((((((.....)))))).(((((((....)))).)))))).))).).))))))))..)).)))))). ( -46.10) >DroSim_CAF1 99960 102 + 1 UCCAUUGCCCGAUGGCCCGU------UCGGUGGCGGACCUCCGGGCAUCGACUGCUUGUGGCCGUCUAAUGACGAGCCGGGCGUCACUCCGCCAUGUCCUUUCGGGCC ......((((((.((..(((------..(((((.(((((((((((((.....)))))).(((((((....)))).)))))).))).).))))))))..)).)))))). ( -46.10) >DroMoj_CAF1 132212 102 + 1 UCCAUUGCCAGCGGGGCCGGCACCGAUGGGCGGU---GCACCAGGUAGCGGCUGCUUGUGGGCAUCGAGUGCUGCGCCGGGCGCCACG---CCAUGCGAUUUUUGGCC ......(((((.((.(((.((((((.....))))---))....(((.(((((.(((((.......))))))))))))).))).)).((---(...)))....))))). ( -45.20) >DroPer_CAF1 94204 99 + 1 GCCGUUGCCGGGUGGUCCAC------UGGGUGGUGGGCCACCAGGCAUCGUCUGUUUGUGCGCGUCUAGCGCUGAGCCAGGCGGCACUCCG---UGCGGCUUCUGGCC (((..((((.((((((((((------(....))))))))))).)))).((((((((((.((((.....)))))))).))))))((((...)---))))))........ ( -53.40) >consensus UCCAUUGCCCGAUGGCCCGU______UCGGUGGCGGACCACCAGGCAUCGACUGCUUGUGGCCGUCUAAUGACGAGCCGGGCGUCACUCCGCCAUGCCCUUUCGGGCC ......((((((....((..........)).((((((.....(((((.....)))))((((.(((((...........)))))))))))))))........)))))). (-26.86 = -25.83 + -1.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:53 2006