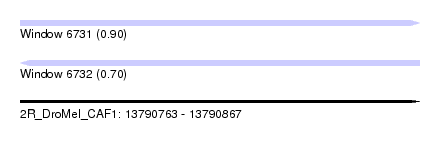
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,790,763 – 13,790,867 |
| Length | 104 |
| Max. P | 0.898152 |

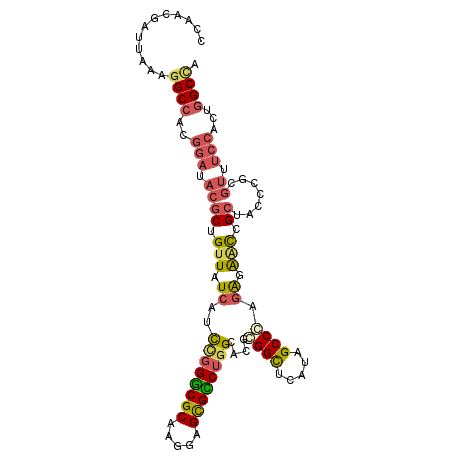
| Location | 13,790,763 – 13,790,867 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -26.81 |
| Energy contribution | -26.98 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
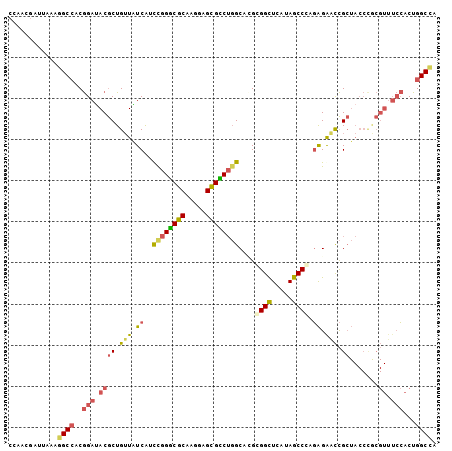
>2R_DroMel_CAF1 13790763 104 + 20766785 CCAACGGUUAAAGGCCUCGGAUACGCUGUUAUCAUUCGGGCGCAAGGAGCGCCUGGCACGCGGCUCAUAGCCGAGAGAACCGCUACCCGCGUUUCCACUGGCCA ............((((..(((.((((.........((((((((.....))))))))...(((((((........)))..)))).....)))).)))...)))). ( -41.20) >DroSec_CAF1 91593 104 + 1 CCAACGAUUAAAGGCCUCGGAUACGCUGUUAUCAUUCGGGCGCAAGGAGUGCCUGGCACGCGGCUCGUAGCCGAGAGAACCGCUACCCGCGUUUCCACUGGCCA ............((((..(((.((((.........((((((((.....))))))))...((((((((....))))....)))).....)))).)))...)))). ( -40.40) >DroSim_CAF1 95453 104 + 1 CCAACGAUUAAAGGCCUCGGAUACGCUGUUAUCAUUCGGGCGCAAGGAGCGCCUGGCACGCGGUUCAUAGCCGAGAGAACCGCUACCCGCGUUUCCACUGGCCA ............((((..(((.((((.........((((((((.....))))))))...(((((((..........))))))).....)))).)))...)))). ( -44.10) >DroEre_CAF1 92002 104 + 1 CCAACGGUUGAAGGCCACGGAAACGCUGUUGUCAUCUGGACGCAAAGAGCGUCUGGCCAGAGGCUCAUAGCCCUGAGGGCCGCUACCUACGUUUCCACUGGCCA ............(((((.(((((((..((.((.....((((((.....))))))((((((.(((.....)))))...)))))).))...)))))))..))))). ( -40.90) >DroYak_CAF1 96271 104 + 1 CCAACGAUUAAACGCAACGGAUACGCUGUUGUCAUCUGGACGCAAAGAGCGUCUGGCCAGAGGCUCAUAGCCCAGAGAGCCGCUACCUACGUUUCCACUGGCAA .............(((((((.....))))))).....((((((.....)))))).(((((.(((((..........)))))((.......)).....))))).. ( -31.20) >DroPer_CAF1 89404 92 + 1 CCAGCCACGAAAGGCCACAGAGACACUGGUCUCGUCCGGUCGCAGGGAGCGGCGUGCCUGGGGCUCAUAGCCCAUGGAAUUG------------GAGCUGGCUA (((((..(((.((((.((.(((((....))))).....(((((.....))))))))))).((((.....))))......)))------------..)))))... ( -39.90) >consensus CCAACGAUUAAAGGCCACGGAUACGCUGUUAUCAUCCGGGCGCAAGGAGCGCCUGGCACGCGGCUCAUAGCCCAGAGAACCGCUACCCGCGUUUCCACUGGCCA ............((((..(((.((((.(((.((..((((((((.....))))))))....((((.....)))).)).))).)).......)).)))...)))). (-26.81 = -26.98 + 0.17)

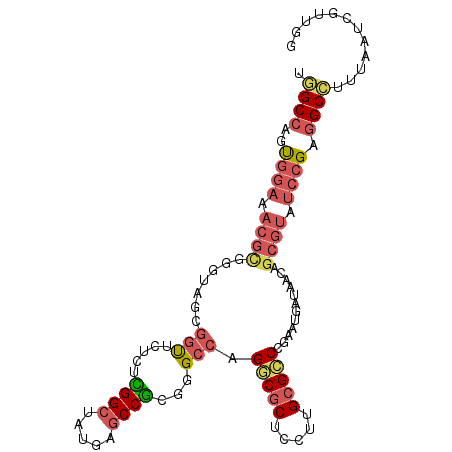
| Location | 13,790,763 – 13,790,867 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -41.03 |
| Consensus MFE | -26.69 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13790763 104 - 20766785 UGGCCAGUGGAAACGCGGGUAGCGGUUCUCUCGGCUAUGAGCCGCGUGCCAGGCGCUCCUUGCGCCCGAAUGAUAACAGCGUAUCCGAGGCCUUUAACCGUUGG .((((..((((.((((.(((((((((((..........))))))).)))).(((((.....)))))............)))).)))).))))............ ( -46.30) >DroSec_CAF1 91593 104 - 1 UGGCCAGUGGAAACGCGGGUAGCGGUUCUCUCGGCUACGAGCCGCGUGCCAGGCACUCCUUGCGCCCGAAUGAUAACAGCGUAUCCGAGGCCUUUAAUCGUUGG .((((..((((.((((((((.(((((((..........)))))))..((.(((....))).))))))...((....)))))).)))).))))............ ( -40.40) >DroSim_CAF1 95453 104 - 1 UGGCCAGUGGAAACGCGGGUAGCGGUUCUCUCGGCUAUGAACCGCGUGCCAGGCGCUCCUUGCGCCCGAAUGAUAACAGCGUAUCCGAGGCCUUUAAUCGUUGG .((((..((((.((((.(((((((((((..........))))))).)))).(((((.....)))))............)))).)))).))))............ ( -46.30) >DroEre_CAF1 92002 104 - 1 UGGCCAGUGGAAACGUAGGUAGCGGCCCUCAGGGCUAUGAGCCUCUGGCCAGACGCUCUUUGCGUCCAGAUGACAACAGCGUUUCCGUGGCCUUCAACCGUUGG .(((((.(((((((((.(.....((((....((((.....))))..)))).(((((.....)))))..........).))))))))))))))............ ( -44.00) >DroYak_CAF1 96271 104 - 1 UUGCCAGUGGAAACGUAGGUAGCGGCUCUCUGGGCUAUGAGCCUCUGGCCAGACGCUCUUUGCGUCCAGAUGACAACAGCGUAUCCGUUGCGUUUAAUCGUUGG (((((..((....))..))))).((((....((((.....))))..)))).(((((.....)))))..(((((....((((((.....))))))...))))).. ( -34.50) >DroPer_CAF1 89404 92 - 1 UAGCCAGCUC------------CAAUUCCAUGGGCUAUGAGCCCCAGGCACGCCGCUCCCUGCGACCGGACGAGACCAGUGUCUCUGUGGCCUUUCGUGGCUGG ...(((((..------------(........((((.....)))).((((.((((((.....))).......(((((....))))).)))))))...)..))))) ( -34.70) >consensus UGGCCAGUGGAAACGCGGGUAGCGGUUCUCUCGGCUAUGAGCCGCGGGCCAGGCGCUCCUUGCGCCCGAAUGAUAACAGCGUAUCCGAGGCCUUUAAUCGUUGG .((((..((((.((((.......(((.....((((.....))))...))).(((((.....)))))............)))).)))).))))............ (-26.69 = -27.38 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:50 2006