
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,780,442 – 13,780,544 |
| Length | 102 |
| Max. P | 0.985997 |

| Location | 13,780,442 – 13,780,544 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -23.51 |
| Energy contribution | -24.27 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13780442 102 + 20766785 UAUGGCUUCCACCUUCUGGAGUCGCAUGUGGAAGCCGCCGCC----GCCGCCUCACAAACGCCUCAGAAUGUUGCUGCUGUAUGGA-GC---AAAAGGAGCAGCAGCAGC- ...((......))(((((..(.((..(((((..((.((....----)).)).)))))..)))..))))).(((((((((((.....-..---.......)))))))))))- ( -32.54) >DroGri_CAF1 95714 105 + 1 UAUGGCUUCCACCUUCUC--GUCGCAUGUGGAAGCCAAUGCCAGCAGCCGCCUCACAAACGCCUGA---UG-CGCUGCUGUACAGCAACGGCAUAAGAAACAACAACAACA ..((((((((((......--.......))))))))))(((((((((((.((.(((........)))---.)-)))))))((......)))))))................. ( -36.82) >DroSec_CAF1 81280 100 + 1 UAUGGCUUCCACCUUCUGGAGUCGCAUGUGGAAGCCGCC-------GCCGCCUCACAAACGCCUCAGAAUGUUGCUGCUGUACGGA-GG---AAAAGGAGCAGCAGCCGCA ...(((((((((....((......)).)))))))))((.-------((.(((((.......((((.....((....))......))-))---.....)))..)).)).)). ( -31.30) >DroSim_CAF1 85207 100 + 1 UAUGGCUUCCACCUUCUGGAGUCGCAUGUGGAAGCCGCC-------GCCGCCUCACAAACGCCUCAGAAUGUUGCUGCUGUACGGA-GG---AAAAGGAGCAGCAGCCGCA ...(((((((((....((......)).)))))))))((.-------((.(((((.......((((.....((....))......))-))---.....)))..)).)).)). ( -31.30) >DroWil_CAF1 99193 81 + 1 UAUGGCUUCCACCUUCUCAAGUCUCAUGUGGAAGCC-------------GCCUCGCAAACGCCUG--AAAUUCGCUGCUGUACGGC---------------AGCAGCAGCA ...(((((((((...............)))))))))-------------((...((....))...--......(((((((.....)---------------)))))).)). ( -31.26) >consensus UAUGGCUUCCACCUUCUGGAGUCGCAUGUGGAAGCCGCC_______GCCGCCUCACAAACGCCUCAGAAUGUUGCUGCUGUACGGA_GC___AAAAGGAGCAGCAGCAGCA ..((((((((((...............))))))))))....................................((((((((..................)))))))).... (-23.51 = -24.27 + 0.76)

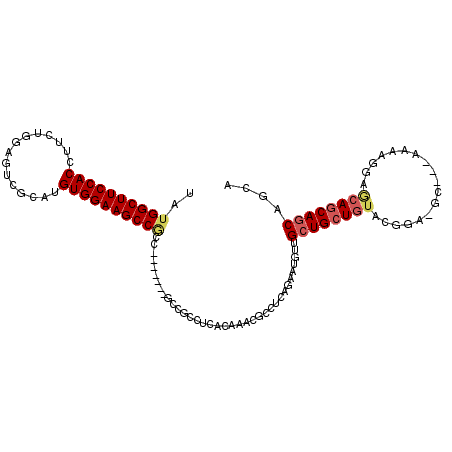
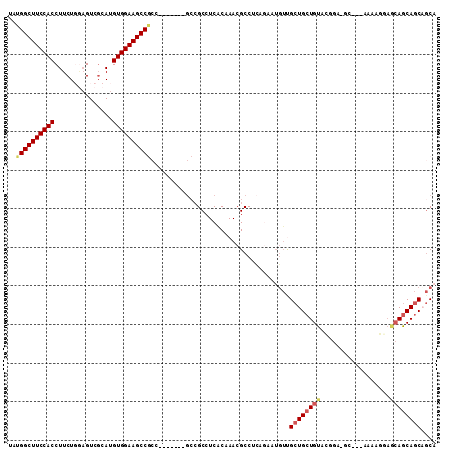
| Location | 13,780,442 – 13,780,544 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -39.63 |
| Consensus MFE | -27.63 |
| Energy contribution | -28.11 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.985997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13780442 102 - 20766785 -GCUGCUGCUGCUCCUUUU---GC-UCCAUACAGCAGCAACAUUCUGAGGCGUUUGUGAGGCGGC----GGCGGCGGCUUCCACAUGCGACUCCAGAAGGUGGAAGCCAUA -(((((((((((..(...(---((-(......))))((..(.....)..))......)..)))))----))))))(((((((((...(.......)...)))))))))... ( -46.50) >DroGri_CAF1 95714 105 - 1 UGUUGUUGUUGUUUCUUAUGCCGUUGCUGUACAGCAGCG-CA---UCAGGCGUUUGUGAGGCGGCUGCUGGCAUUGGCUUCCACAUGCGAC--GAGAAGGUGGAAGCCAUA ((((((.((((((((..(((((..(((((.....)))))-..---...)))))....)))))))).)).)))).((((((((((...(...--..)...)))))))))).. ( -43.00) >DroSec_CAF1 81280 100 - 1 UGCGGCUGCUGCUCCUUUU---CC-UCCGUACAGCAGCAACAUUCUGAGGCGUUUGUGAGGCGGC-------GGCGGCUUCCACAUGCGACUCCAGAAGGUGGAAGCCAUA ....((((((((.......---.(-((((.((.((..((......))..)))).)).))))))))-------)))(((((((((...(.......)...)))))))))... ( -38.20) >DroSim_CAF1 85207 100 - 1 UGCGGCUGCUGCUCCUUUU---CC-UCCGUACAGCAGCAACAUUCUGAGGCGUUUGUGAGGCGGC-------GGCGGCUUCCACAUGCGACUCCAGAAGGUGGAAGCCAUA ....((((((((.......---.(-((((.((.((..((......))..)))).)).))))))))-------)))(((((((((...(.......)...)))))))))... ( -38.20) >DroWil_CAF1 99193 81 - 1 UGCUGCUGCU---------------GCCGUACAGCAGCGAAUUU--CAGGCGUUUGCGAGGC-------------GGCUUCCACAUGAGACUUGAGAAGGUGGAAGCCAUA .(((((((((---------------(.....)))))))......--...((....))..)))-------------(((((((((...............)))))))))... ( -32.26) >consensus UGCUGCUGCUGCUCCUUUU___CC_UCCGUACAGCAGCAACAUUCUGAGGCGUUUGUGAGGCGGC_______GGCGGCUUCCACAUGCGACUCCAGAAGGUGGAAGCCAUA ....(((((((....................)))))))..........(.((((.....)))).)..........(((((((((...(.......)...)))))))))... (-27.63 = -28.11 + 0.48)

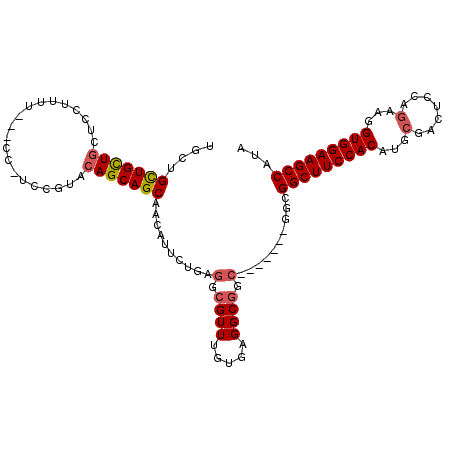
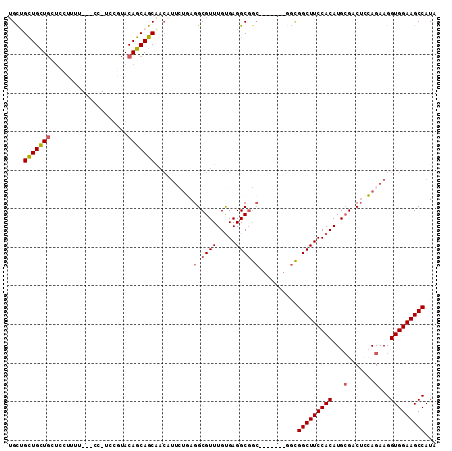
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:48 2006