
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,779,222 – 13,779,326 |
| Length | 104 |
| Max. P | 0.547640 |

| Location | 13,779,222 – 13,779,326 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -17.53 |
| Energy contribution | -17.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13779222 104 - 20766785 UACGAGCAUUGGCGCAAAAGUCCAAGUUCAAAGCCAUAUUUGAGUUGGG--AAAAAUCAAUAAAAAGAUCGCAAGCUGCGAUGGCACAUGCGUGUUAUUUCCAAC-C------------- ...((((.(((((......).))))))))..............((((((--((..............(((((.....)))))(((((....))))).))))))))-.------------- ( -27.40) >DroVir_CAF1 95608 95 - 1 UA------UUGGCGCAAAAGUCCAAGUUCAAAGCCAUAUUUGACUUGGG--AAAAAUCAAUAAAACGAUCGCUUGCUGC---GGCACAUGCGUGUGAGCUGCACU-C------------- ..------..((.(((....(((((((.((((......)))))))))))--...................(((..((((---(.....)))).)..)))))).))-.------------- ( -25.10) >DroGri_CAF1 94425 95 - 1 UA------UUGGCGCAAAAGUCCAAGUUCAAAGCCAUAUUUGACUUGGG--AAAAAUCAAUAAAACGAUCGCUUGCUGC---GGCACAUGCGUGUGUGCCACACU-C------------- ..------...((((((...(((((((.((((......)))))))))))--....(((........)))...)))).))---(((((((....))))))).....-.------------- ( -27.60) >DroWil_CAF1 95865 111 - 1 UA------UUGGCGCAAAAGUCCAAGUUCAAAGCCAUAUUUGACUUGGG--AAAAAUCAAUAAAAAGAUCGCUCGCUGCGAUGGCACAUGCGUGUUAUGCUUAUG-CUUAUUCCCAUUCU ..------.....(((.((((((((((.((((......)))))))))).--................(((((.....)))))(((((....)))))..)))).))-)............. ( -25.70) >DroMoj_CAF1 111383 95 - 1 UA------UUGGCGCAAAAGUCCAAGUUCAAAGCCAUAUUUGACUUGGG--AAAAAUCAAUAAAACGAUCGCUCGCUGG---GGCACAUGCGUGUGAGCCGCACU-C------------- ..------..(((.......(((((((.((((......)))))))))))--.................((((.(((((.---....)).))).))))))).....-.------------- ( -26.70) >DroAna_CAF1 87942 107 - 1 UACGAGCAUGAGCGCAAAAGUCCAAGUUCAAAGCCAUAUUUGAGUUGGGAGAAAAAUCAAUAAAAAGAUCGCAAGCUGCGAUGGCACAUGCGUGUUAUUUCCAUCCC------------- .(((.(((((.((.......(((((.((((((......)))))))))))..................(((((.....))))).)).))))).)))............------------- ( -25.70) >consensus UA______UUGGCGCAAAAGUCCAAGUUCAAAGCCAUAUUUGACUUGGG__AAAAAUCAAUAAAAAGAUCGCUAGCUGC___GGCACAUGCGUGUGAGCCCCACU_C_____________ ...........(((((.....((((((.((((......))))))))))..........................(((.....)))...)))))........................... (-17.53 = -17.87 + 0.33)

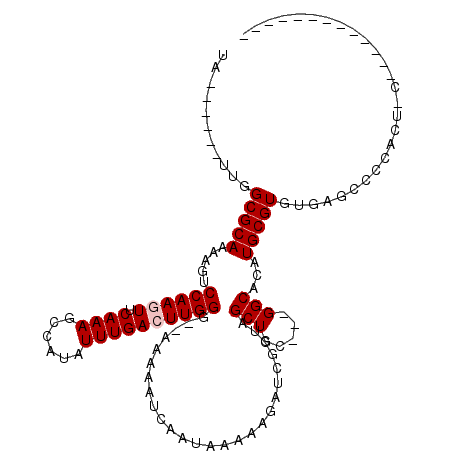
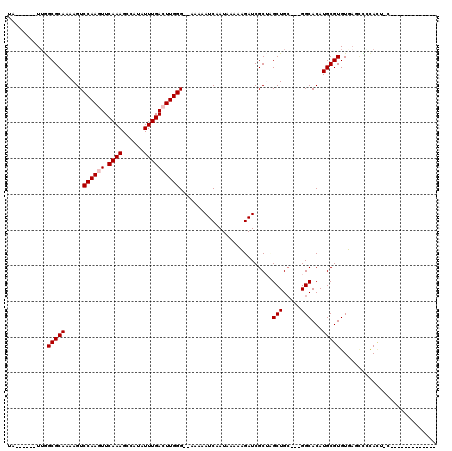
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:46 2006