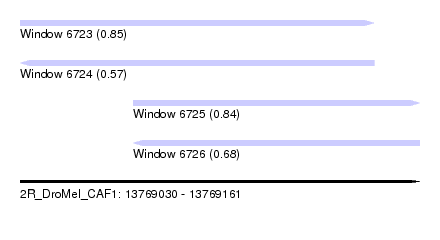
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,769,030 – 13,769,161 |
| Length | 131 |
| Max. P | 0.853751 |

| Location | 13,769,030 – 13,769,146 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
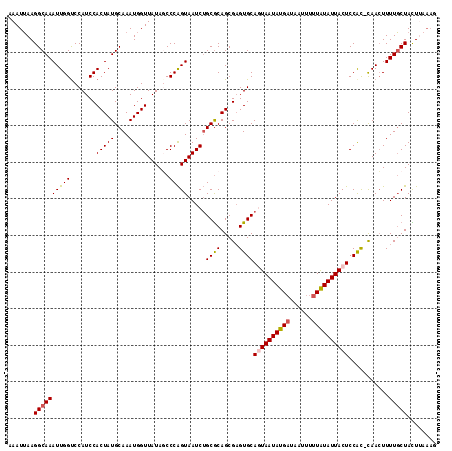
>2R_DroMel_CAF1 13769030 116 + 20766785 AAAUUAAGGCAAAUUGGUCCAUCCACUAUGCAAAUGGUUAUAGCCCAGUAAUCUGCGCAGCGAGUGCAGUAAUAUGAUAAUUUUUAUAUUACUCCAC-CAACUUUUGCUAUUUUAAG .......(((((((((((.....((((.(((....(((....)))(((....))).)))...)))).((((((((((......))))))))))..))-)))..))))))........ ( -28.30) >DroSec_CAF1 70029 117 + 1 AAAUUAAGGCAAAUUGGCCCAUCCACUAUGCAAAUGGUUAUAGCCCAGUAAUCUGCGCAGCGAGUGCAGUAAUAUGAUAAUUUUUAUAUUACUCCAUACAACUUUUGCUACUUAAAG .......(((......)))..........(((((.((((...((.(((....))).))...(.(((.((((((((((......)))))))))).))).))))))))))......... ( -26.30) >DroSim_CAF1 73901 117 + 1 AAAUUAAGGCAAAUUGGUCCAUCCACUAUGCAAAUGGUUAUAGCCCAGUAAUCUGCGCAGCGAGUGCAGUAAUAUGAUAAUUUUUAUAUUACUCCGUACAACUUUUGCUACUUAAAG ...(((((......(((.....)))....(((((.((((...((.(((....))).)).....((((((((((((((......))))))))))..)))))))))))))..))))).. ( -28.10) >DroEre_CAF1 70206 111 + 1 AAAUGAAGGCAAAUUGGUCCAUCCACUAUGCAAAUGGUUAUAUCCCAGUAAUCUGCACAGCGUGUGCAAUAAUAUGUUAAUUUUUGUAUUACUCCGC-UAAAUUUUUCUAUU----- .......(((....(((.....)))..(((((((.((((((......))))))(((((.....)))))..............)))))))......))-).............----- ( -18.10) >DroYak_CAF1 73442 116 + 1 AAAUUAAGGCAAAUUGGUCCAUCCACUAUGCAAAUGGUUAUAUCCCGGUAAUCGGCGCAGCGAGUGCAAUAAUAUGAUAAUUUUUAUAUUACUCCAC-CAACUUUUGCUACUAAAAA .......(((((((((((.....((((.(((...(((((((......)))))))..)))...))))...((((((((......))))))))....))-)))..))))))........ ( -25.80) >consensus AAAUUAAGGCAAAUUGGUCCAUCCACUAUGCAAAUGGUUAUAGCCCAGUAAUCUGCGCAGCGAGUGCAGUAAUAUGAUAAUUUUUAUAUUACUCCAC_CAACUUUUGCUACUUAAAG .......((((((((((.......(((((....)))))......))))).....((((.....))))((((((((((......))))))))))...........)))))........ (-19.58 = -19.90 + 0.32)

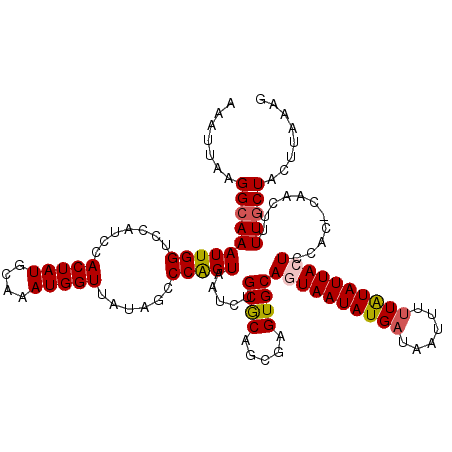
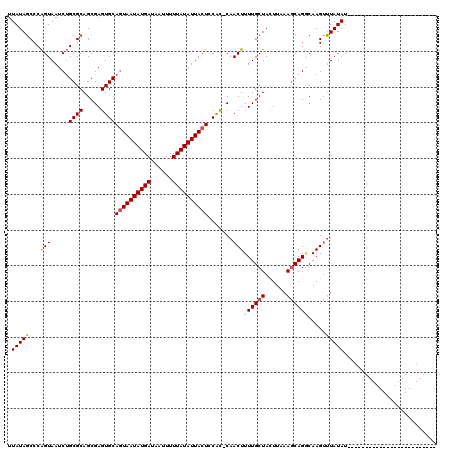
| Location | 13,769,030 – 13,769,146 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13769030 116 - 20766785 CUUAAAAUAGCAAAAGUUG-GUGGAGUAAUAUAAAAAUUAUCAUAUUACUGCACUCGCUGCGCAGAUUACUGGGCUAUAACCAUUUGCAUAGUGGAUGGACCAAUUUGCCUUAAUUU .........((((((((.(-(((.((((((((..........)))))))).)))).)))((.(((....))).)).....(((((..(...)..))))).....)))))........ ( -32.10) >DroSec_CAF1 70029 117 - 1 CUUUAAGUAGCAAAAGUUGUAUGGAGUAAUAUAAAAAUUAUCAUAUUACUGCACUCGCUGCGCAGAUUACUGGGCUAUAACCAUUUGCAUAGUGGAUGGGCCAAUUUGCCUUAAUUU ......(((((...(((.(((((..(((((......)))))))))).)))......)))))((((((...(((.((((..(((((.....))))))))).)))))))))........ ( -28.80) >DroSim_CAF1 73901 117 - 1 CUUUAAGUAGCAAAAGUUGUACGGAGUAAUAUAAAAAUUAUCAUAUUACUGCACUCGCUGCGCAGAUUACUGGGCUAUAACCAUUUGCAUAGUGGAUGGACCAAUUUGCCUUAAUUU ......(((((....(.....)(.((((((((..........)))))))).)....)))))((((((...(((.((((..(((((.....))))))))).)))))))))........ ( -27.20) >DroEre_CAF1 70206 111 - 1 -----AAUAGAAAAAUUUA-GCGGAGUAAUACAAAAAUUAACAUAUUAUUGCACACGCUGUGCAGAUUACUGGGAUAUAACCAUUUGCAUAGUGGAUGGACCAAUUUGCCUUCAUUU -----..............-(((((((((((...............))))))...(((((((((((....(((.......))))))))))))))..........)))))........ ( -22.56) >DroYak_CAF1 73442 116 - 1 UUUUUAGUAGCAAAAGUUG-GUGGAGUAAUAUAAAAAUUAUCAUAUUAUUGCACUCGCUGCGCCGAUUACCGGGAUAUAACCAUUUGCAUAGUGGAUGGACCAAUUUGCCUUAAUUU .........((((((((.(-(((.((((((((..........)))))))).)))).)))...(((.....))).......(((((..(...)..))))).....)))))........ ( -26.90) >consensus CUUUAAGUAGCAAAAGUUG_GUGGAGUAAUAUAAAAAUUAUCAUAUUACUGCACUCGCUGCGCAGAUUACUGGGCUAUAACCAUUUGCAUAGUGGAUGGACCAAUUUGCCUUAAUUU .........(((((..........((((((((..........)))))))).((..(((((.((((((....((.......)))))))).)))))..))......)))))........ (-19.94 = -20.30 + 0.36)

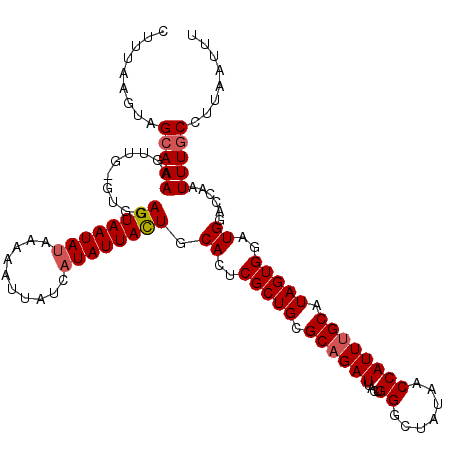
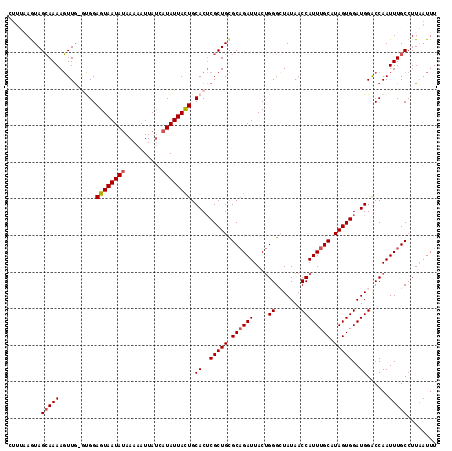
| Location | 13,769,067 – 13,769,161 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13769067 94 + 20766785 UUAUAGCCCAGUAAUCUGCGCAGCGAGUGCAGUAAUAUGAUAAUUUUUAUAUUACUCCAC-CAACUUUUGCUAUUUUAAGCAGCCAAGUUUAUAU------------------------- .....((.(((....))).))...(.(((.((((((((((......)))))))))).)))-)((((((((((......)))))..))))).....------------------------- ( -23.10) >DroSec_CAF1 70066 95 + 1 UUAUAGCCCAGUAAUCUGCGCAGCGAGUGCAGUAAUAUGAUAAUUUUUAUAUUACUCCAUACAACUUUUGCUACUUAAAGCAGGCAAGUUUAUAU------------------------- .....(((..(((...(((((.....)))))(((((((((......)))))))))....)))......((((......)))))))..........------------------------- ( -22.60) >DroSim_CAF1 73938 95 + 1 UUAUAGCCCAGUAAUCUGCGCAGCGAGUGCAGUAAUAUGAUAAUUUUUAUAUUACUCCGUACAACUUUUGCUACUUAAAGCAGGCAAGUUUAUAU------------------------- .....((.(((....))).)).((..((((((((((((((......))))))))))..)))).....(((((......)))))))..........------------------------- ( -23.40) >DroYak_CAF1 73479 119 + 1 UUAUAUCCCGGUAAUCGGCGCAGCGAGUGCAAUAAUAUGAUAAUUUUUAUAUUACUCCAC-CAACUUUUGCUACUAAAAACAGACAAGGAUAUAUGUAUGGUUUGGUAAGACUGAAUGGA .(((((((.(((.....((((.....))))..((((((((......))))))))....))-).....(((((.........)).))))))))))....(.(((..(.....)..))).). ( -21.70) >consensus UUAUAGCCCAGUAAUCUGCGCAGCGAGUGCAGUAAUAUGAUAAUUUUUAUAUUACUCCAC_CAACUUUUGCUACUUAAAGCAGGCAAGUUUAUAU_________________________ .(((((...(((.....((((.....))))((((((((((......)))))))))).......)))((((((......)))))).....))))).......................... (-14.56 = -14.62 + 0.06)

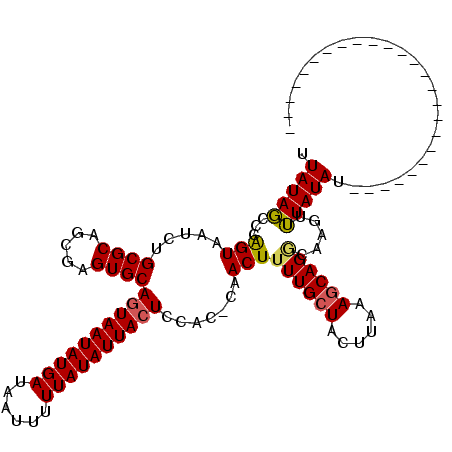
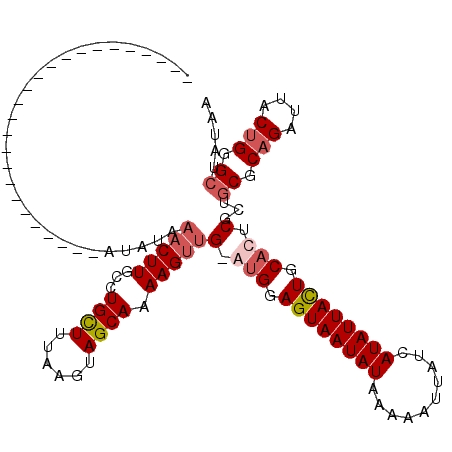
| Location | 13,769,067 – 13,769,161 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -14.77 |
| Energy contribution | -16.15 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13769067 94 - 20766785 -------------------------AUAUAAACUUGGCUGCUUAAAAUAGCAAAAGUUG-GUGGAGUAAUAUAAAAAUUAUCAUAUUACUGCACUCGCUGCGCAGAUUACUGGGCUAUAA -------------------------.............((((......))))..(((.(-(((.((((((((..........)))))))).)))).)))((.(((....))).))..... ( -24.00) >DroSec_CAF1 70066 95 - 1 -------------------------AUAUAAACUUGCCUGCUUUAAGUAGCAAAAGUUGUAUGGAGUAAUAUAAAAAUUAUCAUAUUACUGCACUCGCUGCGCAGAUUACUGGGCUAUAA -------------------------.....(((((...((((......)))).)))))(..((.((((((((..........)))))))).))..)...((.(((....))).))..... ( -20.30) >DroSim_CAF1 73938 95 - 1 -------------------------AUAUAAACUUGCCUGCUUUAAGUAGCAAAAGUUGUACGGAGUAAUAUAAAAAUUAUCAUAUUACUGCACUCGCUGCGCAGAUUACUGGGCUAUAA -------------------------.....(((((...((((......)))).)))))....(.((((((((..........)))))))).).......((.(((....))).))..... ( -19.40) >DroYak_CAF1 73479 119 - 1 UCCAUUCAGUCUUACCAAACCAUACAUAUAUCCUUGUCUGUUUUUAGUAGCAAAAGUUG-GUGGAGUAAUAUAAAAAUUAUCAUAUUAUUGCACUCGCUGCGCCGAUUACCGGGAUAUAA ..........................((((((((.((..(((....(((((.......(-(((.((((((((..........)))))))).)))).)))))...))).)).)))))))). ( -23.80) >consensus _________________________AUAUAAACUUGCCUGCUUUAAGUAGCAAAAGUUG_AUGGAGUAAUAUAAAAAUUAUCAUAUUACUGCACUCGCUGCGCAGAUUACUGGGCUAUAA ..............................(((((...((((......)))).)))))(.(((.((((((((..........)))))))).))).)...((.(((....))).))..... (-14.77 = -16.15 + 1.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:44 2006