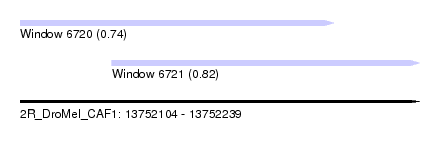
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,752,104 – 13,752,239 |
| Length | 135 |
| Max. P | 0.815193 |

| Location | 13,752,104 – 13,752,210 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -14.85 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
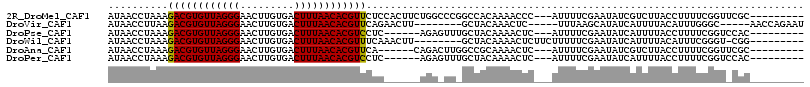
>2R_DroMel_CAF1 13752104 106 + 20766785 ---------CAGCGUUCGGCUAGUUUAAUAAAUAUAUCAAAUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUCUCCACUUCUGGCCCGGCCACAAAACCC---AUUU ---------..(.((..((((((.........(((.....))).......((((((((((((.........))))))))))))........))))))..))).........---.... ( -21.80) >DroVir_CAF1 62041 104 + 1 UUAAUA-AAAAUCGUUAUGCUAGUUUAAUAAAUAUAUCAAAUAACCUUAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUCAGAACUU--------GCUACAAACUC-----UUU ......-......((...((.(((((......(((.....))).......((((((((((((.........))))))))))))..))))).--------)).))......-----... ( -17.20) >DroPse_CAF1 50445 109 + 1 UGCAUGAAAAAUCGUUCAGCUAGUUUAAUAAAUAUAUCAAAUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUCCUC------AGAGUUUGCUACAAAACUC---AUUU .((.((((......))))))..............................((((((((((((.........))))))))))))...------.((((((......))))))---.... ( -24.20) >DroWil_CAF1 55049 110 + 1 UUAAUAAAAAAUCGUUAUGCUAGUUUAAUAAAUAUAUCAAAUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUUCAAACUU--------GCUACAAAACUCUUCUUUU .............((...((.(((((......(((.....)))......(((((((((((((.........))))))))))))).))))).--------)).)).............. ( -17.40) >DroAna_CAF1 57750 109 + 1 UGCAUAAAAAAUUGUUCGGCUAGUUUAAUAAAUAUAUCAAAUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUCA------CAGACUUGGCCGCAAAACUC---AUUU ...........(((..((((((((((......(((.....))).......((((((((((((.........))))))))))))..------.)))).))))))))).....---.... ( -21.60) >DroPer_CAF1 50778 109 + 1 UGCAUGAAAAAUCGUUCAGCUAGUUUAAUAAAUAUAUCAAAUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUCCUC------AGAGUUUGCUACAAAACUC---AUUU .((.((((......))))))..............................((((((((((((.........))))))))))))...------.((((((......))))))---.... ( -24.20) >consensus UGCAUAAAAAAUCGUUCAGCUAGUUUAAUAAAUAUAUCAAAUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUCUC______AGA_UU_GCUACAAAACUC___AUUU .............((..(((..((((.(((....))).))))........((((((((((((.........))))))))))))................))))).............. (-14.85 = -14.60 + -0.25)

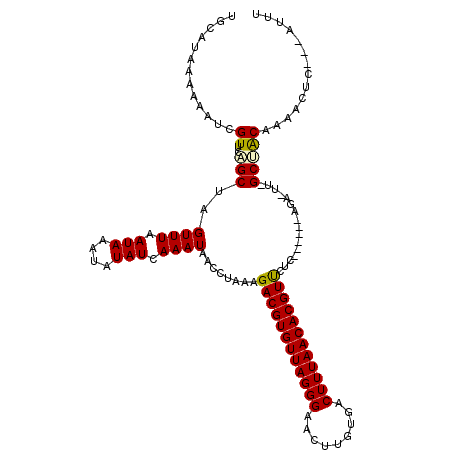
| Location | 13,752,135 – 13,752,239 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -13.35 |
| Energy contribution | -13.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13752135 104 + 20766785 AUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUCUCCACUUCUGGCCCGGCCACAAAACCC---AUUUUCGAAUAUCGUCUUACCUUUUCGGUUCGC--------- ........((((((((((.((((((.(((.......))).)))).)).....((((...))))........---.......)))).))))))(((.....)))....--------- ( -21.40) >DroVir_CAF1 62080 98 + 1 AUAACCUUAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUCAGAACUU--------GCUACAAACUC-----UUUAAGCAUAUCAUUUUACAUUUGGGC-----AACCAGAAU ..........((((((((((((.........))))))))))))..((...(--------(((........-----....))))..))........(((((..-----..))))).. ( -22.30) >DroPse_CAF1 50485 98 + 1 AUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUCCUC------AGAGUUUGCUACAAAACUC---AUUUUCGAAUAUCAUUUUACCUUUUCGGUCCAC--------- ...(((....((((((((((((.........))))))))))))...------.((((((......))))))---..........................)))....--------- ( -23.30) >DroWil_CAF1 55089 98 + 1 AUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUUCAAACUU--------GCUACAAAACUCUUCUUUUUCGAAUAUCAUUUUACAUUUCGGGU-CGG--------- ...(((...(((((((((((((.........))))))))))))).......--------....................((((.............)))))))-...--------- ( -16.42) >DroAna_CAF1 57790 98 + 1 AUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUCA------CAGACUUGGCCGCAAAACUC---AUUUUCGAAUAUCGUCUUACCUUUUCGGUUCGC--------- ..((((.(((((((((((((((.........)))))))))))...------.((((.((..((.((((...---.))))))..))..))))...))))..))))...--------- ( -18.30) >DroPer_CAF1 50818 98 + 1 AUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUCCUC------AGAGUUUGCUACAAAACUC---AUUUUCGAAUAUCAUUUUACCUUUUCGGUCCAC--------- ...(((....((((((((((((.........))))))))))))...------.((((((......))))))---..........................)))....--------- ( -23.30) >consensus AUAACCUAAAGACGUGUUAGGGAACUUGUGACUUUAACACGUUCUC______AGA_UU_GCUACAAAACUC___AUUUUCGAAUAUCAUUUUACCUUUUCGGU_CGC_________ ..........((((((((((((.........))))))))))))......................................................................... (-13.35 = -13.13 + -0.22)
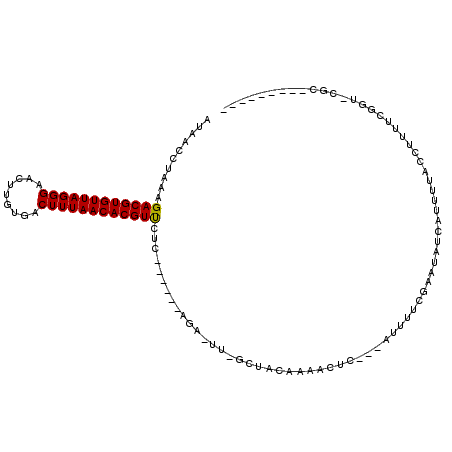
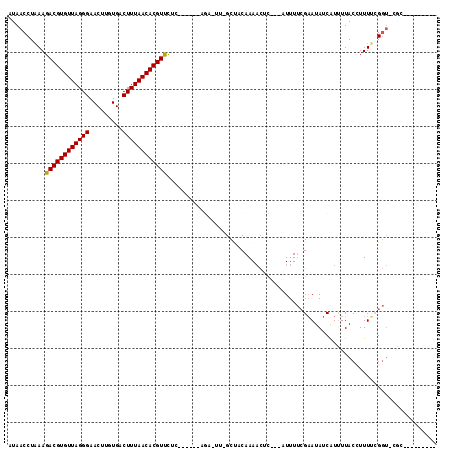
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:40 2006