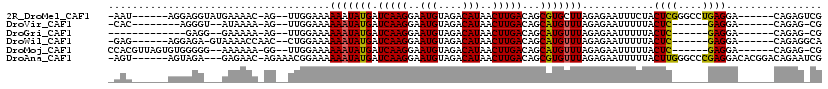
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,744,858 – 13,744,961 |
| Length | 103 |
| Max. P | 0.747751 |

| Location | 13,744,858 – 13,744,961 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -14.83 |
| Consensus MFE | -9.00 |
| Energy contribution | -9.17 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
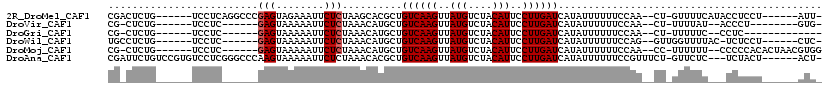
>2R_DroMel_CAF1 13744858 103 + 20766785 CGACUCUG------UCCUCAGGCCCGAGUAGAAAUUCUCUAAGCACGCUGUCAAGUUAUGUCUACAUUCCUUGAUCAUAUUUUUUCCAA--CU-GUUUUCAUACCUCCU------AUU- ......((------....(((((..(..((((.....))))..)..)).((((((..(((....)))..))))))..............--))-)....))........------...- ( -15.00) >DroVir_CAF1 51724 92 + 1 CG-CUCUG------UCCUC------GAGUAAAAAUUCUCUAAACAUGCUGUCAAGUUAUGUCUACAUUCCUUGAUCAUAUUUUUUCCAA--CU-UUUUAU--ACCCU--------GUG- .(-(((..------.....------))))(((((......(((.(((..((((((..(((....)))..))))))..))).))).....--.)-))))..--.....--------...- ( -12.00) >DroGri_CAF1 49978 88 + 1 CG-CUCUG------UCCUC------GAGUAAAAAUUCUCUAAACAUGCUGUCAAGUUAUGUCUACAUUCCUUGAUCAUAUUUUUUCCAA--CU-UUUUUC--CCUC------------- .(-(..((------(....------(((........)))...))).)).((((((..(((....)))..))))))..............--..-......--....------------- ( -11.90) >DroWil_CAF1 48348 97 + 1 UGCCUCUG------UCCUC------GAGUAAAAAUUCUCUAAACAUGCUGUCAAGUUAUGUCUACAUUCCUUGAUCAUAUUUUUUCCAG--GUUGGUUUUAC-UCUCCU------CUC- ........------.....------((((((((..(((..(((.(((..((((((..(((....)))..))))))..))).)))...))--)....))))))-))....------...- ( -18.60) >DroMoj_CAF1 57520 101 + 1 CG-CUCUG------UCCUC------GAGUAAAAAUUCUCUAAACAUGCUGUCAAGUUAUGUCUACAUUCCUUGAUCAUAUUUUUUCCAA--CC-UUUUUU--CCCCCACACUAACGUGG .(-(..((------(....------(((........)))...))).)).((((((..(((....)))..))))))..............--..-......--...((((......)))) ( -15.90) >DroAna_CAF1 48622 108 + 1 CGAUUCUGUCCGUGUCCUCGGGCCCAAGUAAAAAUUCUCUAAACACGCUGUCAAGUUAUGUCUACAUUCCUUGAUCAUAUUUUUUCCGUUUCU-GUUCUC---UCUACU------ACU- .((..(.(((((......)))))..........................((((((..(((....)))..))))))............)..)).-......---......------...- ( -15.60) >consensus CG_CUCUG______UCCUC______GAGUAAAAAUUCUCUAAACAUGCUGUCAAGUUAUGUCUACAUUCCUUGAUCAUAUUUUUUCCAA__CU_GUUUUC___CCCCCU______AUG_ .........................(((........)))..........((((((..(((....)))..))))))............................................ ( -9.00 = -9.17 + 0.17)
| Location | 13,744,858 – 13,744,961 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -13.69 |
| Energy contribution | -13.63 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
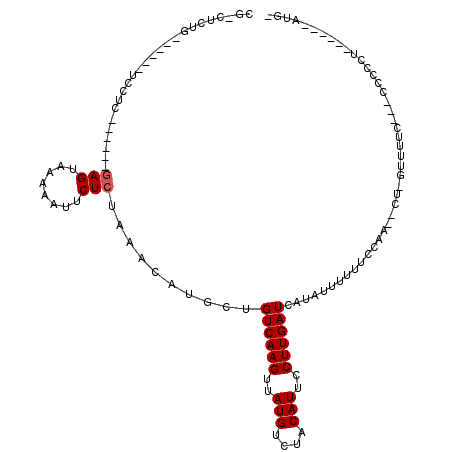
>2R_DroMel_CAF1 13744858 103 - 20766785 -AAU------AGGAGGUAUGAAAAC-AG--UUGGAAAAAAUAUGAUCAAGGAAUGUAGACAUAACUUGACAGCGUGCUUAGAGAAUUUCUACUCGGGCCUGAGGA------CAGAGUCG -...------...(((((((.....-.(--((......))).((.(((((..(((....)))..))))))).))))))).(((........))).((((((....------))).))). ( -19.90) >DroVir_CAF1 51724 92 - 1 -CAC--------AGGGU--AUAAAA-AG--UUGGAAAAAAUAUGAUCAAGGAAUGUAGACAUAACUUGACAGCAUGUUUAGAGAAUUUUUACUC------GAGGA------CAGAG-CG -...--------.((((--(...((-((--((.....(((((((.(((((..(((....)))..)))))...)))))))....)))))))))))------.....------.....-.. ( -17.20) >DroGri_CAF1 49978 88 - 1 -------------GAGG--GAAAAA-AG--UUGGAAAAAAUAUGAUCAAGGAAUGUAGACAUAACUUGACAGCAUGUUUAGAGAAUUUUUACUC------GAGGA------CAGAG-CG -------------(((.--...(((-((--((.....(((((((.(((((..(((....)))..)))))...)))))))....))))))).)))------.....------.....-.. ( -16.60) >DroWil_CAF1 48348 97 - 1 -GAG------AGGAGA-GUAAAACCAAC--CUGGAAAAAAUAUGAUCAAGGAAUGUAGACAUAACUUGACAGCAUGUUUAGAGAAUUUUUACUC------GAGGA------CAGAGGCA -...------....((-((((((((...--..))...(((((((.(((((..(((....)))..)))))...))))))).......))))))))------.....------........ ( -21.80) >DroMoj_CAF1 57520 101 - 1 CCACGUUAGUGUGGGGG--AAAAAA-GG--UUGGAAAAAAUAUGAUCAAGGAAUGUAGACAUAACUUGACAGCAUGUUUAGAGAAUUUUUACUC------GAGGA------CAGAG-CG (((((....)))))...--......-..--............((.(((((..(((....)))..)))))))((.(((((.(((........)))------..)))------))..)-). ( -20.90) >DroAna_CAF1 48622 108 - 1 -AGU------AGUAGA---GAGAAC-AGAAACGGAAAAAAUAUGAUCAAGGAAUGUAGACAUAACUUGACAGCGUGUUUAGAGAAUUUUUACUUGGGCCCGAGGACACGGACAGAAUCG -.((------((((((---((...(-......)....(((((((.(((((..(((....)))..)))))...)))))))......)))))))).....(((......)))))....... ( -18.10) >consensus _AAC______AGGAGG___AAAAAA_AG__UUGGAAAAAAUAUGAUCAAGGAAUGUAGACAUAACUUGACAGCAUGUUUAGAGAAUUUUUACUC______GAGGA______CAGAG_CG .....................................(((((((.(((((..(((....)))..)))))...)))))))............((((....))))................ (-13.69 = -13.63 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:37 2006