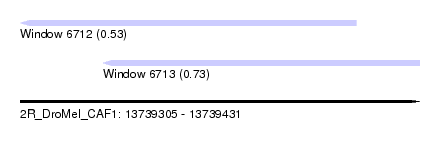
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,739,305 – 13,739,431 |
| Length | 126 |
| Max. P | 0.734549 |

| Location | 13,739,305 – 13,739,411 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
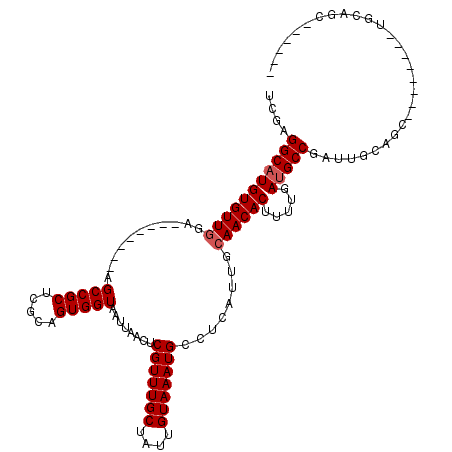
>2R_DroMel_CAF1 13739305 106 - 20766785 UCGGGGCAUGUGUUGGAUCCGUUCGAGCCGCUCGCAGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACACUUUUGAUGCCGAUUGCAAC--------UGCAUC------ ((((.(((.((((((((.....))(((.((.(..(((((((((.........)))))))))..).)).)))....))))))...)).).)))).(((...--------.)))..------ ( -29.60) >DroVir_CAF1 45580 95 - 1 UCGAGGCAUGUGUUG----------AGCCGC-CGUCGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACACUUUUGAUGCCGAUUGCUGC--------UGCUGC------ ..(((((..(.((((----------((((((-....)))))..))))).)((((((....)))))))))))...(((.(((..(((....)))..).)).--------)))...------ ( -27.10) >DroPse_CAF1 37437 106 - 1 UUGGGGCAUGUGUUUGAGCGGCUGGAGCCGCUCGCAGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACACUUUUGAUGCCGAUUGCAGC--------UGCAGC------ .((((((((......(((((((....)))))))((((((((((.........))))))))))..))))))))..(((.((....)).)))...((((...--------.)))).------ ( -40.40) >DroGri_CAF1 44549 95 - 1 UCGAGGCAUGUGUUG----------AGCCGC-CGUCGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACACUUUUGAUGCCGAUUGCUGC--------UGCUGC------ ..(((((..(.((((----------((((((-....)))))..))))).)((((((....)))))))))))...(((.(((..(((....)))..).)).--------)))...------ ( -27.10) >DroWil_CAF1 41577 100 - 1 UCGAGGCAUGUGUUGGAGCU------GCCGCGCACAGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACACUUUUGAUGCCGUUUGCAGC--------CACAGC------ ..(((((((((((.((....------.))))))((((((((((.........))))))))))..))))))).((((((.((..........)))))))).--------......------ ( -29.60) >DroAna_CAF1 42689 111 - 1 UCGAGGCAUGUGUUGAU---------GCCGCUCGCCGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACACUUUUCAAGCCGAUUGCUGCUUGCAAGUUGCAACUGGAAG ..(((((..(.((((((---------(((((.....))))).)))))).)((((((....))))))))))).(((((((...((.(((((.(....).))))).)))))))))....... ( -36.90) >consensus UCGAGGCAUGUGUUGGA________AGCCGCUCGCAGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACACUUUUGAUGCCGAUUGCAGC________UGCAGC______ ....(((((((((((...........(((((.....)))))........(((((((....)))))))........)))))).....)))))............................. (-22.28 = -22.62 + 0.33)

| Location | 13,739,331 – 13,739,431 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.50 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -18.69 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13739331 100 - 20766785 ------------GC--UGG-GC-----CUCCUCUAUAAGGUCGGGGCAUGUGUUGGAUCCGUUCGAGCCGCUCGCAGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACA ------------((--..(-((-----((........)))))((((((((((((.((.....)).)).)))..((((((((((.........))))))))))..)))))))...)).... ( -31.40) >DroVir_CAF1 45606 86 - 1 ------------GC--AGU-G--------GCUCUAUAAGGUCGAGGCAUGUGUUG----------AGCCGC-CGUCGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACA ------------((--(((-(--------(((......))))(((((..(.((((----------((((((-....)))))..))))).)((((((....)))))))))))))))).... ( -30.20) >DroGri_CAF1 44575 86 - 1 ------------AC--UGU-G--------GCUCUAUAAGGUCGAGGCAUGUGUUG----------AGCCGC-CGUCGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACA ------------..--(((-(--------(((......))))(((((..(.((((----------((((((-....)))))..))))).)((((((....)))))))))))...)))... ( -25.90) >DroWil_CAF1 41603 106 - 1 AGCAGCAACAACGA--AGA-GC-----CUUCUCUAUAAGGUCGAGGCAUGUGUUGGAGCU------GCCGCGCACAGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACA ....((((......--.((-.(-----(((......))))))(((((((((((.((....------.))))))((((((((((.........))))))))))..))))))).)))).... ( -33.60) >DroMoj_CAF1 50593 87 - 1 ------------GC--AGUUG--------CCUCUAUAAGGUCGAGGCAUGUGUUG----------AGCCGA-CGUCGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACA ------------((--(((.(--------(((.....)))).(((((((......----------(((.((-((..((........)).)))).))).......)))))))))))).... ( -27.32) >DroAna_CAF1 42729 98 - 1 ------------ACAUUGG-GCAUUGGCUCCUCUAUAAGGUCGAGGCAUGUGUUGAU---------GCCGCUCGCCGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACA ------------.......-(((..((((.........))))(((((..(.((((((---------(((((.....))))).)))))).)((((((....)))))))))))..))).... ( -29.60) >consensus ____________GC__AGU_G________CCUCUAUAAGGUCGAGGCAUGUGUUG__________AGCCGC_CGCCGUGGUAAUUAACUCGUUUGCUAUUGUAAAUGCCUCAUUGCAACA ..........................................(((((...................(((((.....))))).........((((((....)))))))))))......... (-18.69 = -18.72 + 0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:32 2006